Chapter 28 Logistic regression with nominal response
library(nnet)
library(tidyverse)28.1 Finding non-missing values
* This is to prepare you for something in the next question. It’s meant to be easy.
In R, the code NA stands for “missing value” or
“value not known”. In R, NA should not have quotes around
it. (It is a special code, not a piece of text.)
Create a vector
vthat contains some numbers and some missing values, usingc(). Put those values into a one-column data frame.Obtain a new column containing
is.na(v). When is this true and when is this false?The symbol
!means “not” in R (and other programming languages). What does!is.na(v)do? Create a new column containing that.Use
filterto display just the rows of your data frame that have a non-missing value ofv.
28.3 Alligator food
What do alligators most like to eat? 219 alligators were captured in four Florida lakes. Each alligator’s stomach contents were observed, and the food that the alligator had eaten was classified into one of five categories: fish, invertebrates (such as snails or insects), reptiles (such as turtles), birds, and “other” (such as amphibians, plants or rocks). The researcher noted for each alligator what that alligator had most of in its stomach, as well as the gender of each alligator and whether it was “large” or “small” (greater or less than 2.3 metres in length). The data can be found in link. The numbers in the data set (apart from the first column) are all frequencies. (You can ignore that first column “profile”.)
Our aim is to predict food type from the other variables.
Read in the data and display the first few lines. Describe how the data are not “tidy”.
Use
pivot_longerto arrange the data suitably for analysis (which will be usingmultinom). Demonstrate (by looking at the first few rows of your new data frame) that you now have something tidy.What is different about this problem, compared to Question here, that would make
multinomthe right tool to use?Fit a suitable multinomial model predicting food type from gender, size and lake. Does each row represent one alligator or more than one? If more than one, account for this in your modelling.
Do a test to see whether
Gendershould stay in the model. (This will entail fitting another model.) What do you conclude?Predict the probability that an alligator prefers each food type, given its size, gender (if necessary) and the lake it was found in, using the more appropriate of the two models that you have fitted so far. This means (i) making a data frame for prediction, and (ii) obtaining and displaying the predicted probabilities in a way that is easy to read.
What do you think is the most important way in which the lakes differ? (Hint: look at where the biggest predicted probabilities are.)
How would you describe the major difference between the diets of the small and large alligators?
28.4 Crimes in San Francisco
The data in link is a subset of a huge dataset of crimes committed in San Francisco between 2003 and 2015. The variables are:
Dates: the date and time of the crimeCategory: the category of crime, eg. “larceny” or “vandalism” (response).Descript: detailed description of crime.DayOfWeek: the day of the week of the crime.PdDistrict: the name of the San Francisco Police Department district in which the crime took place.Resolution: how the crime was resolvedAddress: approximate street address of crimeX: longitudeY: latitude
Our aim is to see whether the category of crime depends on the day of the week and the district in which it occurred. However, there are a lot of crime categories, so we will focus on the top four “interesting” ones, which are the ones included in this data file.
Some of the model-fitting takes a while (you’ll see why below). If
you’re using R Markdown, you can wait for the models to fit each time
you re-run your document, or insert cache=T in the top line
of your code chunk (the one with r in curly brackets in it,
above the actual code). Put a comma and the cache=T inside
the curly brackets. What that does is to re-run that code chunk only
if it changes; if it hasn’t changed it will use the saved results from
last time it was run. That can save you a lot of waiting around.
Read in the data and display the dataset (or, at least, part of it).
Fit a multinomial logistic regression that predicts crime category from day of week and district. (You don’t need to look at it.) The model-fitting produces some output. (If you’re using R Markdown, that will come with it.)
Fit a model that predicts Category from only the district. Hand in the output from the fitting process as well.
Use
anovato compare the two models you just obtained. What does theanovatell you?Using your preferred model, obtain predicted probabilities that a crime will be of each of these four categories for each day of the week in the
TENDERLOINdistrict (the name is ALL CAPS). This will mean constructing a data frame to predict from, obtaining the predictions and then displaying them suitably.Describe briefly how the weekend days Saturday and Sunday differ from the rest.
28.5 Crimes in San Francisco – the data
The data in link is a huge dataset of crimes committed in San Francisco between 2003 and 2015. The variables are:
Dates: the date and time of the crimeCategory: the category of crime, eg. “larceny” or “vandalism” (response).Descript: detailed description of crime.DayOfWeek: the day of the week of the crime.PdDistrict: the name of the San Francisco Police Department district in which the crime took place.Resolution: how the crime was resolvedAddress: approximate street address of crimeX: longitudeY: latitude
Our aim is to see whether the category of crime depends on the day of the week and the district in which it occurred. However, there are a lot of crime categories, so we will focus on the top four “interesting” ones, which we will have to discover.
Read in the data and verify that you have these columns and a lot of rows. (The data may take a moment to read in. You will see why.)
How is the response variable here different to the one in the question about steak preferences (and therefore why would
multinomfrom packagennetbe the method of choice)?Find out which crime categories there are, and arrange them in order of how many crimes there were in each category.
Which are the four most frequent “interesting” crime categories, that is to say, not including “other offenses” and “non-criminal”? Get them into a vector called
my.crimes. See if you can find a way of doing this that doesn’t involve typing them in (for full marks).(Digression, but needed for the next part.) The R vector
letterscontains the lowercase letters fromatoz. Consider the vector('a','m',3,'Q'). Some of these are found amongst the lowercase letters, and some not. Type these into a vectorvand explain briefly whyv %in% lettersproduces what it does.We are going to
filteronly the rows of our data frame that have one of the crimes inmy.crimesas theirCategory. Also,selectonly the columnsCategory,DayOfWeekandPdDistrict. Save the resulting data frame and display its structure. (You should have a lot fewer rows than you did before.)Save these data in a file
sfcrime1.csv.
28.6 What sports do these athletes play?
The data at link are physical and physiological measurements of 202 male and female Australian elite athletes. The data values are separated by tabs. We are going to see whether we can predict the sport an athlete plays from their height and weight.
The sports, if you care, are respectively basketball, “field athletics” (eg. shot put, javelin throw, long jump etc.), gymnastics, netball, rowing, swimming, 400m running, tennis, sprinting (100m or 200m running), water polo.
Read in the data and display the first few rows.
Make a scatterplot of height vs. weight, with the points coloured by what sport the athlete plays. Put height on the \(x\)-axis and weight on the \(y\)-axis.
Explain briefly why a multinomial model (
multinomfromnnet) would be the best thing to use to predict sport played from the other variables.Fit a suitable model for predicting sport played from height and weight. (You don’t need to look at the results.) 100 steps isn’t quite enough, so set
maxitequal to a larger number to allow the estimation to finish.Demonstrate using
anovathatWtshould not be removed from this model.Make a data frame consisting of all combinations of
Ht160, 180 and 200 (cm), andWt50, 75, and 100 (kg), and use it to obtain predicted probabilities of athletes of those heights and weights playing each of the sports. Display the results. You might have to display them smaller, or reduce the number of decimal places36 to fit them on the page.For an athlete who is 180 cm tall and weighs 100 kg, what sport would you guess they play? How sure are you that you are right? Explain briefly.
My solutions follow:
28.7 Finding non-missing values
* This is to prepare you for something in the next question. It’s meant to be easy.
In R, the code NA stands for “missing value” or
“value not known”. In R, NA should not have quotes around
it. (It is a special code, not a piece of text.)
- Create a vector
vthat contains some numbers and some missing values, usingc(). Put those values into a one-column data frame.
Solution
Like this. The arrangement of numbers and missing values doesn’t matter, as long as you have some of each:
v <- c(1, 2, NA, 4, 5, 6, 9, NA, 11)
mydata <- tibble(v)
mydata## # A tibble: 9 × 1
## v
## <dbl>
## 1 1
## 2 2
## 3 NA
## 4 4
## 5 5
## 6 6
## 7 9
## 8 NA
## 9 11This has one column called v.
\(\blacksquare\)
- Obtain a new column containing
is.na(v). When is this true and when is this false?
Solution
mydata <- mydata %>% mutate(isna = is.na(v))
mydata## # A tibble: 9 × 2
## v isna
## <dbl> <lgl>
## 1 1 FALSE
## 2 2 FALSE
## 3 NA TRUE
## 4 4 FALSE
## 5 5 FALSE
## 6 6 FALSE
## 7 9 FALSE
## 8 NA TRUE
## 9 11 FALSEThis is TRUE if the corresponding element of v is
missing (in my case, the third value and the second-last one), and
FALSE otherwise (when there is an actual value there).
\(\blacksquare\)
- The symbol
!means “not” in R (and other programming languages). What does!is.na(v)do? Create a new column containing that.
Solution
Try it and see. Give it whatever name you like. My name reflects that I know what it’s going to do:
mydata <- mydata %>% mutate(notisna = !is.na(v))
mydata## # A tibble: 9 × 3
## v isna notisna
## <dbl> <lgl> <lgl>
## 1 1 FALSE TRUE
## 2 2 FALSE TRUE
## 3 NA TRUE FALSE
## 4 4 FALSE TRUE
## 5 5 FALSE TRUE
## 6 6 FALSE TRUE
## 7 9 FALSE TRUE
## 8 NA TRUE FALSE
## 9 11 FALSE TRUEThis is the logical opposite of is.na: it’s true if there is
a value, and false if it’s missing.
\(\blacksquare\)
- Use
filterto display just the rows of your data frame that have a non-missing value ofv.
Solution
filter takes a column to say which rows to pick, in
which case the column should contain something that either is
TRUE or FALSE, or something that can be
interpreted that way:
mydata %>% filter(notisna)## # A tibble: 7 × 3
## v isna notisna
## <dbl> <lgl> <lgl>
## 1 1 FALSE TRUE
## 2 2 FALSE TRUE
## 3 4 FALSE TRUE
## 4 5 FALSE TRUE
## 5 6 FALSE TRUE
## 6 9 FALSE TRUE
## 7 11 FALSE TRUEor you can provide filter something that can be calculated
from what’s in the data frame, and also returns something that is
either true or false:
mydata %>% filter(!is.na(v))## # A tibble: 7 × 3
## v isna notisna
## <dbl> <lgl> <lgl>
## 1 1 FALSE TRUE
## 2 2 FALSE TRUE
## 3 4 FALSE TRUE
## 4 5 FALSE TRUE
## 5 6 FALSE TRUE
## 6 9 FALSE TRUE
## 7 11 FALSE TRUEIn either case, I only have non-missing values of v.
\(\blacksquare\)
28.9 Alligator food
What do alligators most like to eat? 219 alligators were captured in four Florida lakes. Each alligator’s stomach contents were observed, and the food that the alligator had eaten was classified into one of five categories: fish, invertebrates (such as snails or insects), reptiles (such as turtles), birds, and “other” (such as amphibians, plants or rocks). The researcher noted for each alligator what that alligator had most of in its stomach, as well as the gender of each alligator and whether it was “large” or “small” (greater or less than 2.3 metres in length). The data can be found in link. The numbers in the data set (apart from the first column) are all frequencies. (You can ignore that first column “profile”.)
Our aim is to predict food type from the other variables.
- Read in the data and display the first few lines. Describe how the data are not “tidy”.
Solution
Separated by exactly one space:
my_url <- "http://ritsokiguess.site/datafiles/alligator.txt"
gators.orig <- read_delim(my_url, " ")## Rows: 16 Columns: 9
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: " "
## chr (3): Gender, Size, Lake
## dbl (6): profile, Fish, Invertebrate, Reptile, Bird, Other
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.gators.orig## # A tibble: 16 × 9
## profile Gender Size Lake Fish Invertebrate Reptile Bird Other
## <dbl> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 f <2.3 george 3 9 1 0 1
## 2 2 m <2.3 george 13 10 0 2 2
## 3 3 f >2.3 george 8 1 0 0 1
## 4 4 m >2.3 george 9 0 0 1 2
## 5 5 f <2.3 hancock 16 3 2 2 3
## 6 6 m <2.3 hancock 7 1 0 0 5
## 7 7 f >2.3 hancock 3 0 1 2 3
## 8 8 m >2.3 hancock 4 0 0 1 2
## 9 9 f <2.3 oklawaha 3 9 1 0 2
## 10 10 m <2.3 oklawaha 2 2 0 0 1
## 11 11 f >2.3 oklawaha 0 1 0 1 0
## 12 12 m >2.3 oklawaha 13 7 6 0 0
## 13 13 f <2.3 trafford 2 4 1 1 4
## 14 14 m <2.3 trafford 3 7 1 0 1
## 15 15 f >2.3 trafford 0 1 0 0 0
## 16 16 m >2.3 trafford 8 6 6 3 5The last five columns are all frequencies. Or, one of the variables (food type) is spread over five columns instead of being contained in one. Either is good.
My choice of “temporary” name reflects that I’m going to obtain a
“tidy” data frame called gators in a moment.
\(\blacksquare\)
- Use
pivot_longerto arrange the data suitably for analysis (which will be usingmultinom). Demonstrate (by looking at the first few rows of your new data frame) that you now have something tidy.
Solution
I’m creating my “official” data frame here:
gators.orig %>%
pivot_longer(Fish:Other, names_to = "Food.type", values_to = "Frequency") -> gators
gators## # A tibble: 80 × 6
## profile Gender Size Lake Food.type Frequency
## <dbl> <chr> <chr> <chr> <chr> <dbl>
## 1 1 f <2.3 george Fish 3
## 2 1 f <2.3 george Invertebrate 9
## 3 1 f <2.3 george Reptile 1
## 4 1 f <2.3 george Bird 0
## 5 1 f <2.3 george Other 1
## 6 2 m <2.3 george Fish 13
## 7 2 m <2.3 george Invertebrate 10
## 8 2 m <2.3 george Reptile 0
## 9 2 m <2.3 george Bird 2
## 10 2 m <2.3 george Other 2
## # … with 70 more rowsI gave my column names Capital Letters to make them consistent with the others (and in an attempt to stop infesting my brain with annoying variable-name errors when I fit models later).
Looking at the first few lines reveals that I now have a column of food types and one column of frequencies, both of which are what I wanted. I can check that I have all the different food types by finding the distinct ones:
gators %>% distinct(Food.type)## # A tibble: 5 × 1
## Food.type
## <chr>
## 1 Fish
## 2 Invertebrate
## 3 Reptile
## 4 Bird
## 5 Other(Think about why count would be confusing here.)
Note that Food.type is text (chr) rather than being a
factor. I’ll hold my breath and see what happens when I fit a model
where it is supposed to be a factor.
\(\blacksquare\)
- What is different about this problem, compared to
Question here, that would make
multinomthe right tool to use?
Solution
Look at the response variable Food.type (or whatever
you called it): this has multiple categories, but they are
not ordered in any logical way. Thus, in short, a nominal
response.
\(\blacksquare\)
- Fit a suitable multinomial model predicting food type from gender, size and lake. Does each row represent one alligator or more than one? If more than one, account for this in your modelling.
Solution
Each row of the tidy gators represents as many
alligators as are in the Frequency column. That is, if
you look at female small alligators in Lake George that ate
mainly fish, there are three of those.42
This to remind you to include the weights piece,
otherwise multinom will assume that you have one
observation per line and not as many as the number in
Frequency.
That is the
reason that count earlier would have been confusing:
it would have told you how many rows contained each
food type, rather than how many alligators, and these
would have been different:
gators %>% count(Food.type)## # A tibble: 5 × 2
## Food.type n
## <chr> <int>
## 1 Bird 16
## 2 Fish 16
## 3 Invertebrate 16
## 4 Other 16
## 5 Reptile 16gators %>% count(Food.type, wt = Frequency)## # A tibble: 5 × 2
## Food.type n
## <chr> <dbl>
## 1 Bird 13
## 2 Fish 94
## 3 Invertebrate 61
## 4 Other 32
## 5 Reptile 19Each food type appears on 16 rows, but is the favoured diet of very
different numbers of alligators. Note the use of wt=
to specify a frequency variable.43
You ought to understand why those are different.
All right, back to modelling:
library(nnet)
gators.1 <- multinom(Food.type ~ Gender + Size + Lake,
weights = Frequency, data = gators
)## # weights: 35 (24 variable)
## initial value 352.466903
## iter 10 value 270.228588
## iter 20 value 268.944257
## final value 268.932741
## convergedThis worked, even though Food.type was actually text. I guess
it got converted to a factor. The ordering of the levels doesn’t
matter here anyway, since this is not an ordinal model.
No need to look at it, since the output is kind of confusing anyway:
summary(gators.1)## Call:
## multinom(formula = Food.type ~ Gender + Size + Lake, data = gators,
## weights = Frequency)
##
## Coefficients:
## (Intercept) Genderm Size>2.3 Lakehancock Lakeoklawaha
## Fish 2.4322304 0.60674971 -0.7308535 -0.5751295 0.5513785
## Invertebrate 2.6012531 0.14378459 -2.0671545 -2.3557377 1.4645820
## Other 1.0014505 0.35423803 -1.0214847 0.1914537 0.5775317
## Reptile -0.9829064 -0.02053375 -0.1741207 0.5534169 3.0807416
## Laketrafford
## Fish -1.23681053
## Invertebrate -0.08096493
## Other 0.32097943
## Reptile 1.82333205
##
## Std. Errors:
## (Intercept) Genderm Size>2.3 Lakehancock Lakeoklawaha
## Fish 0.7706940 0.6888904 0.6523273 0.7952147 1.210229
## Invertebrate 0.7917210 0.7292510 0.7084028 0.9463640 1.232835
## Other 0.8747773 0.7623738 0.7250455 0.9072182 1.374545
## Reptile 1.2827234 0.9088217 0.8555051 1.3797755 1.591542
## Laketrafford
## Fish 0.8661187
## Invertebrate 0.8814625
## Other 0.9589807
## Reptile 1.3388017
##
## Residual Deviance: 537.8655
## AIC: 585.8655You get one coefficient for each variable (along the top) and for each response group (down the side), using the first group as a baseline everywhere. These numbers are hard to interpret; doing predictions is much easier.
\(\blacksquare\)
- Do a test to see whether
Gendershould stay in the model. (This will entail fitting another model.) What do you conclude?
Solution
The other model to fit is the one without the variable you’re testing:
gators.2 <- update(gators.1, . ~ . - Gender)## # weights: 30 (20 variable)
## initial value 352.466903
## iter 10 value 272.246275
## iter 20 value 270.046891
## final value 270.040139
## convergedI did update here to show you that it works, but of course
there’s no problem in just writing out the whole model again and
taking out Gender, preferably by copying and pasting:
gators.2x <- multinom(Food.type ~ Size + Lake,
weights = Frequency, data = gators
)## # weights: 30 (20 variable)
## initial value 352.466903
## iter 10 value 272.246275
## iter 20 value 270.046891
## final value 270.040139
## convergedand then you compare the models with and without Gender using anova:
anova(gators.2, gators.1)## Likelihood ratio tests of Multinomial Models
##
## Response: Food.type
## Model Resid. df Resid. Dev Test Df LR stat. Pr(Chi)
## 1 Size + Lake 300 540.0803
## 2 Gender + Size + Lake 296 537.8655 1 vs 2 4 2.214796 0.6963214The P-value is not small, so the two models fit equally well, and
therefore we should go with the smaller, simpler one: that is, the one
without Gender.
Sometimes drop1 works here too (and sometimes it doesn’t, for
reasons I haven’t figured out):
drop1(gators.1, test = "Chisq")## trying - Gender## Error in if (trace) {: argument is not interpretable as logicalI don’t even know what this error message means, never mind what to do about it.
\(\blacksquare\)
- Predict the probability that an alligator prefers each food type, given its size, gender (if necessary) and the lake it was found in, using the more appropriate of the two models that you have fitted so far. This means (i) making a data frame for prediction, and (ii) obtaining and displaying the predicted probabilities in a way that is easy to read.
Solution
Our best model gators.2 contains size and lake, so we need to predict for all combinations of those.
You might think of this first, which is easiest:
predictions(gators.2, variables = c("Size", "Lake"), type = "probs")## rowid type group predicted std.error Food.type Size Lake
## 1 1 probs Bird 0.029671502 0.026490803 Fish <2.3 george
## 2 2 probs Bird 0.029671502 0.026490803 Fish <2.3 george
## 3 3 probs Bird 0.029671502 0.026490803 Fish <2.3 george
## 4 4 probs Bird 0.029671502 0.026490803 Fish <2.3 george
## 5 5 probs Bird 0.070400215 0.014591121 Fish <2.3 hancock
## 6 6 probs Bird 0.070400215 0.014591121 Fish <2.3 hancock
## 7 7 probs Bird 0.070400215 0.014591121 Fish <2.3 hancock
## 8 8 probs Bird 0.070400215 0.014591121 Fish <2.3 hancock
## 9 9 probs Bird 0.008818267 0.031423530 Fish <2.3 oklawaha
## 10 10 probs Bird 0.008818267 0.031423530 Fish <2.3 oklawaha
## 11 11 probs Bird 0.008818267 0.031423530 Fish <2.3 oklawaha
## 12 12 probs Bird 0.008818267 0.031423530 Fish <2.3 oklawaha
## 13 13 probs Bird 0.035892547 0.030455498 Fish <2.3 trafford
## 14 14 probs Bird 0.035892547 0.030455498 Fish <2.3 trafford
## 15 15 probs Bird 0.035892547 0.030455498 Fish <2.3 trafford
## 16 16 probs Bird 0.035892547 0.030455498 Fish <2.3 trafford
## 17 17 probs Bird 0.081071082 0.028409610 Fish >2.3 george
## 18 18 probs Bird 0.081071082 0.028409610 Fish >2.3 george
## 19 19 probs Bird 0.081071082 0.028409610 Fish >2.3 george
## 20 20 probs Bird 0.081071082 0.028409610 Fish >2.3 george
## 21 21 probs Bird 0.140898571 0.033231369 Fish >2.3 hancock
## 22 22 probs Bird 0.140898571 0.033231369 Fish >2.3 hancock
## 23 23 probs Bird 0.140898571 0.033231369 Fish >2.3 hancock
## 24 24 probs Bird 0.140898571 0.033231369 Fish >2.3 hancock
## 25 25 probs Bird 0.029419560 0.043739196 Fish >2.3 oklawaha
## 26 26 probs Bird 0.029419560 0.043739196 Fish >2.3 oklawaha
## 27 27 probs Bird 0.029419560 0.043739196 Fish >2.3 oklawaha
## 28 28 probs Bird 0.029419560 0.043739196 Fish >2.3 oklawaha
## 29 29 probs Bird 0.108222209 0.049665725 Fish >2.3 trafford
## 30 30 probs Bird 0.108222209 0.049665725 Fish >2.3 trafford
## 31 31 probs Bird 0.108222209 0.049665725 Fish >2.3 trafford
## 32 32 probs Bird 0.108222209 0.049665725 Fish >2.3 trafford
## 33 1 probs Fish 0.452103200 0.190911359 Fish <2.3 george
## 34 2 probs Fish 0.452103200 0.190911359 Fish <2.3 george
## 35 3 probs Fish 0.452103200 0.190911359 Fish <2.3 george
## 36 4 probs Fish 0.452103200 0.190911359 Fish <2.3 george
## 37 5 probs Fish 0.535303986 0.053136442 Fish <2.3 hancock
## 38 6 probs Fish 0.535303986 0.053136442 Fish <2.3 hancock
## 39 7 probs Fish 0.535303986 0.053136442 Fish <2.3 hancock
## 40 8 probs Fish 0.535303986 0.053136442 Fish <2.3 hancock
## 41 9 probs Fish 0.258187239 0.137187952 Fish <2.3 oklawaha
## 42 10 probs Fish 0.258187239 0.137187952 Fish <2.3 oklawaha
## 43 11 probs Fish 0.258187239 0.137187952 Fish <2.3 oklawaha
## 44 12 probs Fish 0.258187239 0.137187952 Fish <2.3 oklawaha
## 45 13 probs Fish 0.184299727 0.500835571 Fish <2.3 trafford
## 46 14 probs Fish 0.184299727 0.500835571 Fish <2.3 trafford
## 47 15 probs Fish 0.184299727 0.500835571 Fish <2.3 trafford
## 48 16 probs Fish 0.184299727 0.500835571 Fish <2.3 trafford
## 49 17 probs Fish 0.657439424 0.251626520 Fish >2.3 george
## 50 18 probs Fish 0.657439424 0.251626520 Fish >2.3 george
## 51 19 probs Fish 0.657439424 0.251626520 Fish >2.3 george
## 52 20 probs Fish 0.657439424 0.251626520 Fish >2.3 george
## 53 21 probs Fish 0.570196832 0.288825624 Fish >2.3 hancock
## 54 22 probs Fish 0.570196832 0.288825624 Fish >2.3 hancock
## 55 23 probs Fish 0.570196832 0.288825624 Fish >2.3 hancock
## 56 24 probs Fish 0.570196832 0.288825624 Fish >2.3 hancock
## 57 25 probs Fish 0.458436758 0.314296880 Fish >2.3 oklawaha
## 58 26 probs Fish 0.458436758 0.314296880 Fish >2.3 oklawaha
## 59 27 probs Fish 0.458436758 0.314296880 Fish >2.3 oklawaha
## 60 28 probs Fish 0.458436758 0.314296880 Fish >2.3 oklawaha
## 61 29 probs Fish 0.295752571 0.220261657 Fish >2.3 trafford
## 62 30 probs Fish 0.295752571 0.220261657 Fish >2.3 trafford
## 63 31 probs Fish 0.295752571 0.220261657 Fish >2.3 trafford
## 64 32 probs Fish 0.295752571 0.220261657 Fish >2.3 trafford
## 65 1 probs Invertebrate 0.412856987 0.028347945 Fish <2.3 george
## 66 2 probs Invertebrate 0.412856987 0.028347945 Fish <2.3 george
## 67 3 probs Invertebrate 0.412856987 0.028347945 Fish <2.3 george
## 68 4 probs Invertebrate 0.412856987 0.028347945 Fish <2.3 george
## 69 5 probs Invertebrate 0.093098852 0.011544508 Fish <2.3 hancock
## 70 6 probs Invertebrate 0.093098852 0.011544508 Fish <2.3 hancock
## 71 7 probs Invertebrate 0.093098852 0.011544508 Fish <2.3 hancock
## 72 8 probs Invertebrate 0.093098852 0.011544508 Fish <2.3 hancock
## 73 9 probs Invertebrate 0.601895180 0.072899406 Fish <2.3 oklawaha
## 74 10 probs Invertebrate 0.601895180 0.072899406 Fish <2.3 oklawaha
## 75 11 probs Invertebrate 0.601895180 0.072899406 Fish <2.3 oklawaha
## 76 12 probs Invertebrate 0.601895180 0.072899406 Fish <2.3 oklawaha
## 77 13 probs Invertebrate 0.516837696 0.031040421 Fish <2.3 trafford
## 78 14 probs Invertebrate 0.516837696 0.031040421 Fish <2.3 trafford
## 79 15 probs Invertebrate 0.516837696 0.031040421 Fish <2.3 trafford
## 80 16 probs Invertebrate 0.516837696 0.031040421 Fish <2.3 trafford
## 81 17 probs Invertebrate 0.139678770 0.266063328 Fish >2.3 george
## 82 18 probs Invertebrate 0.139678770 0.266063328 Fish >2.3 george
## 83 19 probs Invertebrate 0.139678770 0.266063328 Fish >2.3 george
## 84 20 probs Invertebrate 0.139678770 0.266063328 Fish >2.3 george
## 85 21 probs Invertebrate 0.023071788 0.253432814 Fish >2.3 hancock
## 86 22 probs Invertebrate 0.023071788 0.253432814 Fish >2.3 hancock
## 87 23 probs Invertebrate 0.023071788 0.253432814 Fish >2.3 hancock
## 88 24 probs Invertebrate 0.023071788 0.253432814 Fish >2.3 hancock
## 89 25 probs Invertebrate 0.248644081 0.340363100 Fish >2.3 oklawaha
## 90 26 probs Invertebrate 0.248644081 0.340363100 Fish >2.3 oklawaha
## 91 27 probs Invertebrate 0.248644081 0.340363100 Fish >2.3 oklawaha
## 92 28 probs Invertebrate 0.248644081 0.340363100 Fish >2.3 oklawaha
## 93 29 probs Invertebrate 0.192961478 0.293280651 Fish >2.3 trafford
## 94 30 probs Invertebrate 0.192961478 0.293280651 Fish >2.3 trafford
## 95 31 probs Invertebrate 0.192961478 0.293280651 Fish >2.3 trafford
## 96 32 probs Invertebrate 0.192961478 0.293280651 Fish >2.3 trafford
## 97 1 probs Other 0.093801903 0.030785528 Fish <2.3 george
## 98 2 probs Other 0.093801903 0.030785528 Fish <2.3 george
## 99 3 probs Other 0.093801903 0.030785528 Fish <2.3 george
## 100 4 probs Other 0.093801903 0.030785528 Fish <2.3 george
## 101 5 probs Other 0.253741634 0.025649228 Fish <2.3 hancock
## 102 6 probs Other 0.253741634 0.025649228 Fish <2.3 hancock
## 103 7 probs Other 0.253741634 0.025649228 Fish <2.3 hancock
## 104 8 probs Other 0.253741634 0.025649228 Fish <2.3 hancock
## 105 9 probs Other 0.053872406 0.048740534 Fish <2.3 oklawaha
## 106 10 probs Other 0.053872406 0.048740534 Fish <2.3 oklawaha
## 107 11 probs Other 0.053872406 0.048740534 Fish <2.3 oklawaha
## 108 12 probs Other 0.053872406 0.048740534 Fish <2.3 oklawaha
## 109 13 probs Other 0.174203303 0.287237268 Fish <2.3 trafford
## 110 14 probs Other 0.174203303 0.287237268 Fish <2.3 trafford
## 111 15 probs Other 0.174203303 0.287237268 Fish <2.3 trafford
## 112 16 probs Other 0.174203303 0.287237268 Fish <2.3 trafford
## 113 17 probs Other 0.097911930 0.005970406 Fish >2.3 george
## 114 18 probs Other 0.097911930 0.005970406 Fish >2.3 george
## 115 19 probs Other 0.097911930 0.005970406 Fish >2.3 george
## 116 20 probs Other 0.097911930 0.005970406 Fish >2.3 george
## 117 21 probs Other 0.194008990 0.008762261 Fish >2.3 hancock
## 118 22 probs Other 0.194008990 0.008762261 Fish >2.3 hancock
## 119 23 probs Other 0.194008990 0.008762261 Fish >2.3 hancock
## 120 24 probs Other 0.194008990 0.008762261 Fish >2.3 hancock
## 121 25 probs Other 0.068662061 0.007119469 Fish >2.3 oklawaha
## 122 26 probs Other 0.068662061 0.007119469 Fish >2.3 oklawaha
## 123 27 probs Other 0.068662061 0.007119469 Fish >2.3 oklawaha
## 124 28 probs Other 0.068662061 0.007119469 Fish >2.3 oklawaha
## 125 29 probs Other 0.200662408 0.106641288 Fish >2.3 trafford
## 126 30 probs Other 0.200662408 0.106641288 Fish >2.3 trafford
## 127 31 probs Other 0.200662408 0.106641288 Fish >2.3 trafford
## 128 32 probs Other 0.200662408 0.106641288 Fish >2.3 trafford
## 129 1 probs Reptile 0.011566408 0.137747240 Fish <2.3 george
## 130 2 probs Reptile 0.011566408 0.137747240 Fish <2.3 george
## 131 3 probs Reptile 0.011566408 0.137747240 Fish <2.3 george
## 132 4 probs Reptile 0.011566408 0.137747240 Fish <2.3 george
## 133 5 probs Reptile 0.047455313 0.011939732 Fish <2.3 hancock
## 134 6 probs Reptile 0.047455313 0.011939732 Fish <2.3 hancock
## 135 7 probs Reptile 0.047455313 0.011939732 Fish <2.3 hancock
## 136 8 probs Reptile 0.047455313 0.011939732 Fish <2.3 hancock
## 137 9 probs Reptile 0.077226908 0.055750872 Fish <2.3 oklawaha
## 138 10 probs Reptile 0.077226908 0.055750872 Fish <2.3 oklawaha
## 139 11 probs Reptile 0.077226908 0.055750872 Fish <2.3 oklawaha
## 140 12 probs Reptile 0.077226908 0.055750872 Fish <2.3 oklawaha
## 141 13 probs Reptile 0.088766727 0.321998572 Fish <2.3 trafford
## 142 14 probs Reptile 0.088766727 0.321998572 Fish <2.3 trafford
## 143 15 probs Reptile 0.088766727 0.321998572 Fish <2.3 trafford
## 144 16 probs Reptile 0.088766727 0.321998572 Fish <2.3 trafford
## 145 17 probs Reptile 0.023898795 0.025910578 Fish >2.3 george
## 146 18 probs Reptile 0.023898795 0.025910578 Fish >2.3 george
## 147 19 probs Reptile 0.023898795 0.025910578 Fish >2.3 george
## 148 20 probs Reptile 0.023898795 0.025910578 Fish >2.3 george
## 149 21 probs Reptile 0.071823819 0.003294197 Fish >2.3 hancock
## 150 22 probs Reptile 0.071823819 0.003294197 Fish >2.3 hancock
## 151 23 probs Reptile 0.071823819 0.003294197 Fish >2.3 hancock
## 152 24 probs Reptile 0.071823819 0.003294197 Fish >2.3 hancock
## 153 25 probs Reptile 0.194837540 0.005228529 Fish >2.3 oklawaha
## 154 26 probs Reptile 0.194837540 0.005228529 Fish >2.3 oklawaha
## 155 27 probs Reptile 0.194837540 0.005228529 Fish >2.3 oklawaha
## 156 28 probs Reptile 0.194837540 0.005228529 Fish >2.3 oklawaha
## 157 29 probs Reptile 0.202401335 0.119901577 Fish >2.3 trafford
## 158 30 probs Reptile 0.202401335 0.119901577 Fish >2.3 trafford
## 159 31 probs Reptile 0.202401335 0.119901577 Fish >2.3 trafford
## 160 32 probs Reptile 0.202401335 0.119901577 Fish >2.3 trafford
## Frequency
## 1 0.0
## 2 1.0
## 3 3.5
## 4 16.0
## 5 0.0
## 6 1.0
## 7 3.5
## 8 16.0
## 9 0.0
## 10 1.0
## 11 3.5
## 12 16.0
## 13 0.0
## 14 1.0
## 15 3.5
## 16 16.0
## 17 0.0
## 18 1.0
## 19 3.5
## 20 16.0
## 21 0.0
## 22 1.0
## 23 3.5
## 24 16.0
## 25 0.0
## 26 1.0
## 27 3.5
## 28 16.0
## 29 0.0
## 30 1.0
## 31 3.5
## 32 16.0
## 33 0.0
## 34 1.0
## 35 3.5
## 36 16.0
## 37 0.0
## 38 1.0
## 39 3.5
## 40 16.0
## 41 0.0
## 42 1.0
## 43 3.5
## 44 16.0
## 45 0.0
## 46 1.0
## 47 3.5
## 48 16.0
## 49 0.0
## 50 1.0
## 51 3.5
## 52 16.0
## 53 0.0
## 54 1.0
## 55 3.5
## 56 16.0
## 57 0.0
## 58 1.0
## 59 3.5
## 60 16.0
## 61 0.0
## 62 1.0
## 63 3.5
## 64 16.0
## 65 0.0
## 66 1.0
## 67 3.5
## 68 16.0
## 69 0.0
## 70 1.0
## 71 3.5
## 72 16.0
## 73 0.0
## 74 1.0
## 75 3.5
## 76 16.0
## 77 0.0
## 78 1.0
## 79 3.5
## 80 16.0
## 81 0.0
## 82 1.0
## 83 3.5
## 84 16.0
## 85 0.0
## 86 1.0
## 87 3.5
## 88 16.0
## 89 0.0
## 90 1.0
## 91 3.5
## 92 16.0
## 93 0.0
## 94 1.0
## 95 3.5
## 96 16.0
## 97 0.0
## 98 1.0
## 99 3.5
## 100 16.0
## 101 0.0
## 102 1.0
## 103 3.5
## 104 16.0
## 105 0.0
## 106 1.0
## 107 3.5
## 108 16.0
## 109 0.0
## 110 1.0
## 111 3.5
## 112 16.0
## 113 0.0
## 114 1.0
## 115 3.5
## 116 16.0
## 117 0.0
## 118 1.0
## 119 3.5
## 120 16.0
## 121 0.0
## 122 1.0
## 123 3.5
## 124 16.0
## 125 0.0
## 126 1.0
## 127 3.5
## 128 16.0
## 129 0.0
## 130 1.0
## 131 3.5
## 132 16.0
## 133 0.0
## 134 1.0
## 135 3.5
## 136 16.0
## 137 0.0
## 138 1.0
## 139 3.5
## 140 16.0
## 141 0.0
## 142 1.0
## 143 3.5
## 144 16.0
## 145 0.0
## 146 1.0
## 147 3.5
## 148 16.0
## 149 0.0
## 150 1.0
## 151 3.5
## 152 16.0
## 153 0.0
## 154 1.0
## 155 3.5
## 156 16.0
## 157 0.0
## 158 1.0
## 159 3.5
## 160 16.0except that this uses the original dataframe, in which the sizes and lakes are repeated (once for each food type, of which there are five):
gators## # A tibble: 80 × 6
## profile Gender Size Lake Food.type Frequency
## <dbl> <chr> <chr> <chr> <chr> <dbl>
## 1 1 f <2.3 george Fish 3
## 2 1 f <2.3 george Invertebrate 9
## 3 1 f <2.3 george Reptile 1
## 4 1 f <2.3 george Bird 0
## 5 1 f <2.3 george Other 1
## 6 2 m <2.3 george Fish 13
## 7 2 m <2.3 george Invertebrate 10
## 8 2 m <2.3 george Reptile 0
## 9 2 m <2.3 george Bird 2
## 10 2 m <2.3 george Other 2
## # … with 70 more rowsYou can persist with the above, if you are careful:
predictions(gators.2, variables = c("Size", "Lake"), type = "probs") %>%
select(group, predicted, Size, Lake) %>%
pivot_wider(names_from = group, values_from = predicted)## Warning: Values from `predicted` are not uniquely identified; output will contain list-cols.
## * Use `values_fn = list` to suppress this warning.
## * Use `values_fn = {summary_fun}` to summarise duplicates.
## * Use the following dplyr code to identify duplicates.
## {data} %>%
## dplyr::group_by(Size, Lake, group) %>%
## dplyr::summarise(n = dplyr::n(), .groups = "drop") %>%
## dplyr::filter(n > 1L)## # A tibble: 8 × 7
## Size Lake Bird Fish Invertebrate Other Reptile
## <chr> <chr> <list> <list> <list> <list> <list>
## 1 <2.3 george <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 2 <2.3 hancock <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 3 <2.3 oklawaha <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 4 <2.3 trafford <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 5 >2.3 george <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 6 >2.3 hancock <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 7 >2.3 oklawaha <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>
## 8 >2.3 trafford <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]> <dbl [4]>and now you have four numbers in each cell (that are actually all the same), corresponding to the four repeats of size and lake in the original dataset.
A little reading of the help for pivot_wider reveals that there is an option values_fn. This is for exactly this case, where pivoting wider has resulted in multiple values per cell. The input to values_fn is the name of a function that will be used to summarize the four values in each cell. Since those four values are all the same in each case (they are predictions for the same values of size and lake), you could summarize them by any of mean, median, max, min, etc:44
predictions(gators.2, variables = c("Size", "Lake"), type = "probs") %>%
select(group, predicted, Size, Lake) %>%
pivot_wider(names_from = group, values_from = predicted, values_fn = mean) -> preds1
preds1## # A tibble: 8 × 7
## Size Lake Bird Fish Invertebrate Other Reptile
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 <2.3 george 0.0297 0.452 0.413 0.0938 0.0116
## 2 <2.3 hancock 0.0704 0.535 0.0931 0.254 0.0475
## 3 <2.3 oklawaha 0.00882 0.258 0.602 0.0539 0.0772
## 4 <2.3 trafford 0.0359 0.184 0.517 0.174 0.0888
## 5 >2.3 george 0.0811 0.657 0.140 0.0979 0.0239
## 6 >2.3 hancock 0.141 0.570 0.0231 0.194 0.0718
## 7 >2.3 oklawaha 0.0294 0.458 0.249 0.0687 0.195
## 8 >2.3 trafford 0.108 0.296 0.193 0.201 0.202So that works, but you might not have thought of all that.
The other way is to get hold of the different names of lakes and sizes, for which this is one way, and then use datagrid:
Lakes <- gators %>% distinct(Lake) %>% pull(Lake)
Lakes## [1] "george" "hancock" "oklawaha" "trafford"Sizes <- gators %>% distinct(Size) %>% pull(Size)
Sizes## [1] "<2.3" ">2.3"I didn’t need to think about Genders because that’s not in
the better model. See below for what happens if you include it
anyway.
I have persisted with the Capital Letters, for consistency.
Then:
new <- datagrid(model = gators.2, Size = Sizes, Lake = Lakes)
new## Frequency Size Lake
## 1: 2.7375 <2.3 george
## 2: 2.7375 <2.3 hancock
## 3: 2.7375 <2.3 oklawaha
## 4: 2.7375 <2.3 trafford
## 5: 2.7375 >2.3 george
## 6: 2.7375 >2.3 hancock
## 7: 2.7375 >2.3 oklawaha
## 8: 2.7375 >2.3 traffordThe Frequency column is the mean Frequency observed, which was (kinda) part of the model, but won’t affect the predictions that we get.
Next:
predictions(gators.2, newdata = new, type = "probs")## rowid type group predicted std.error Frequency Size Lake
## 1 1 probs Bird 0.029671502 0.026490803 2.7375 <2.3 george
## 2 2 probs Bird 0.070400215 0.014591121 2.7375 <2.3 hancock
## 3 3 probs Bird 0.008818267 0.031423530 2.7375 <2.3 oklawaha
## 4 4 probs Bird 0.035892547 0.030455498 2.7375 <2.3 trafford
## 5 5 probs Bird 0.081071082 0.028409610 2.7375 >2.3 george
## 6 6 probs Bird 0.140898571 0.033231369 2.7375 >2.3 hancock
## 7 7 probs Bird 0.029419560 0.043739196 2.7375 >2.3 oklawaha
## 8 8 probs Bird 0.108222209 0.049665725 2.7375 >2.3 trafford
## 9 1 probs Fish 0.452103200 0.190911359 2.7375 <2.3 george
## 10 2 probs Fish 0.535303986 0.053136442 2.7375 <2.3 hancock
## 11 3 probs Fish 0.258187239 0.137187952 2.7375 <2.3 oklawaha
## 12 4 probs Fish 0.184299727 0.500835571 2.7375 <2.3 trafford
## 13 5 probs Fish 0.657439424 0.251626520 2.7375 >2.3 george
## 14 6 probs Fish 0.570196832 0.288825624 2.7375 >2.3 hancock
## 15 7 probs Fish 0.458436758 0.314296880 2.7375 >2.3 oklawaha
## 16 8 probs Fish 0.295752571 0.220261657 2.7375 >2.3 trafford
## 17 1 probs Invertebrate 0.412856987 0.028347945 2.7375 <2.3 george
## 18 2 probs Invertebrate 0.093098852 0.011544508 2.7375 <2.3 hancock
## 19 3 probs Invertebrate 0.601895180 0.072899406 2.7375 <2.3 oklawaha
## 20 4 probs Invertebrate 0.516837696 0.031040421 2.7375 <2.3 trafford
## 21 5 probs Invertebrate 0.139678770 0.266063328 2.7375 >2.3 george
## 22 6 probs Invertebrate 0.023071788 0.253432814 2.7375 >2.3 hancock
## 23 7 probs Invertebrate 0.248644081 0.340363100 2.7375 >2.3 oklawaha
## 24 8 probs Invertebrate 0.192961478 0.293280651 2.7375 >2.3 trafford
## 25 1 probs Other 0.093801903 0.030785528 2.7375 <2.3 george
## 26 2 probs Other 0.253741634 0.025649228 2.7375 <2.3 hancock
## 27 3 probs Other 0.053872406 0.048740534 2.7375 <2.3 oklawaha
## 28 4 probs Other 0.174203303 0.287237268 2.7375 <2.3 trafford
## 29 5 probs Other 0.097911930 0.005970406 2.7375 >2.3 george
## 30 6 probs Other 0.194008990 0.008762261 2.7375 >2.3 hancock
## 31 7 probs Other 0.068662061 0.007119469 2.7375 >2.3 oklawaha
## 32 8 probs Other 0.200662408 0.106641288 2.7375 >2.3 trafford
## 33 1 probs Reptile 0.011566408 0.137747240 2.7375 <2.3 george
## 34 2 probs Reptile 0.047455313 0.011939732 2.7375 <2.3 hancock
## 35 3 probs Reptile 0.077226908 0.055750872 2.7375 <2.3 oklawaha
## 36 4 probs Reptile 0.088766727 0.321998572 2.7375 <2.3 trafford
## 37 5 probs Reptile 0.023898795 0.025910578 2.7375 >2.3 george
## 38 6 probs Reptile 0.071823819 0.003294197 2.7375 >2.3 hancock
## 39 7 probs Reptile 0.194837540 0.005228529 2.7375 >2.3 oklawaha
## 40 8 probs Reptile 0.202401335 0.119901577 2.7375 >2.3 traffordThis is, you’ll remember, long, with one row for each diet (in group), so we need to select the columns of interest to us, and then pivot wider:
predictions(gators.2, newdata = new, type = "probs") %>%
select(rowid, group, predicted, Size, Lake) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 8 × 8
## rowid Size Lake Bird Fish Invertebrate Other Reptile
## <int> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 <2.3 george 0.0297 0.452 0.413 0.0938 0.0116
## 2 2 <2.3 hancock 0.0704 0.535 0.0931 0.254 0.0475
## 3 3 <2.3 oklawaha 0.00882 0.258 0.602 0.0539 0.0772
## 4 4 <2.3 trafford 0.0359 0.184 0.517 0.174 0.0888
## 5 5 >2.3 george 0.0811 0.657 0.140 0.0979 0.0239
## 6 6 >2.3 hancock 0.141 0.570 0.0231 0.194 0.0718
## 7 7 >2.3 oklawaha 0.0294 0.458 0.249 0.0687 0.195
## 8 8 >2.3 trafford 0.108 0.296 0.193 0.201 0.202Either of these two ways is good to get to this point, or some other way that also gets you here.
If you thought that the better model was the one with Gender
in it, or you otherwise forgot that you didn’t need Gender
then you needed to do something like this as well:
Genders <- gators %>% distinct(Gender) %>% pull(Gender)
new <- datagrid(model = gators.2, Lake = Lakes, Size = Sizes, Gender = Genders)## Warning: Some of the variable names are missing from the model data: Gendernew## Frequency Lake Size Gender
## 1: 2.7375 george <2.3 f
## 2: 2.7375 george <2.3 m
## 3: 2.7375 george >2.3 f
## 4: 2.7375 george >2.3 m
## 5: 2.7375 hancock <2.3 f
## 6: 2.7375 hancock <2.3 m
## 7: 2.7375 hancock >2.3 f
## 8: 2.7375 hancock >2.3 m
## 9: 2.7375 oklawaha <2.3 f
## 10: 2.7375 oklawaha <2.3 m
## 11: 2.7375 oklawaha >2.3 f
## 12: 2.7375 oklawaha >2.3 m
## 13: 2.7375 trafford <2.3 f
## 14: 2.7375 trafford <2.3 m
## 15: 2.7375 trafford >2.3 f
## 16: 2.7375 trafford >2.3 mIf you predict this in the model without Gender, you’ll get
the following:
predictions(gators.2, newdata = new, type = "probs") %>%
select(rowid, group, predicted, Lake, Size, Gender) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 16 × 9
## rowid Lake Size Gender Bird Fish Invertebrate Other Reptile
## <int> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 george <2.3 f 0.0297 0.452 0.413 0.0938 0.0116
## 2 2 george <2.3 m 0.0297 0.452 0.413 0.0938 0.0116
## 3 3 george >2.3 f 0.0811 0.657 0.140 0.0979 0.0239
## 4 4 george >2.3 m 0.0811 0.657 0.140 0.0979 0.0239
## 5 5 hancock <2.3 f 0.0704 0.535 0.0931 0.254 0.0475
## 6 6 hancock <2.3 m 0.0704 0.535 0.0931 0.254 0.0475
## 7 7 hancock >2.3 f 0.141 0.570 0.0231 0.194 0.0718
## 8 8 hancock >2.3 m 0.141 0.570 0.0231 0.194 0.0718
## 9 9 oklawaha <2.3 f 0.00882 0.258 0.602 0.0539 0.0772
## 10 10 oklawaha <2.3 m 0.00882 0.258 0.602 0.0539 0.0772
## 11 11 oklawaha >2.3 f 0.0294 0.458 0.249 0.0687 0.195
## 12 12 oklawaha >2.3 m 0.0294 0.458 0.249 0.0687 0.195
## 13 13 trafford <2.3 f 0.0359 0.184 0.517 0.174 0.0888
## 14 14 trafford <2.3 m 0.0359 0.184 0.517 0.174 0.0888
## 15 15 trafford >2.3 f 0.108 0.296 0.193 0.201 0.202
## 16 16 trafford >2.3 m 0.108 0.296 0.193 0.201 0.202Here, the predictions for each gender are exactly the same,
because not having Gender in the model means that we take its
effect to be exactly zero.
Alternatively, if you really thought the model with Gender was the
better one, then you’d do this:
predictions(gators.1, newdata = new, type = "probs") %>%
select(rowid, group, predicted, Lake, Size, Gender) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 16 × 9
## rowid Lake Size Gender Bird Fish Invertebrate Other Reptile
## <int> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 george <2.3 f 0.0345 0.393 0.465 0.0940 0.0129
## 2 2 george <2.3 m 0.0240 0.501 0.373 0.0930 0.00879
## 3 3 george >2.3 f 0.105 0.578 0.180 0.103 0.0332
## 4 4 george >2.3 m 0.0679 0.683 0.134 0.0948 0.0209
## 5 5 hancock <2.3 f 0.0792 0.507 0.101 0.261 0.0515
## 6 6 hancock <2.3 m 0.0511 0.601 0.0755 0.240 0.0326
## 7 7 hancock >2.3 f 0.167 0.516 0.0271 0.199 0.0914
## 8 8 hancock >2.3 m 0.110 0.624 0.0206 0.186 0.0591
## 9 9 oklawaha <2.3 f 0.0109 0.215 0.633 0.0527 0.0885
## 10 10 oklawaha <2.3 m 0.00837 0.303 0.564 0.0579 0.0668
## 11 11 oklawaha >2.3 f 0.0378 0.359 0.279 0.0659 0.258
## 12 12 oklawaha >2.3 m 0.0276 0.483 0.236 0.0688 0.185
## 13 13 trafford <2.3 f 0.0438 0.145 0.545 0.165 0.102
## 14 14 trafford <2.3 m 0.0344 0.209 0.494 0.184 0.0782
## 15 15 trafford >2.3 f 0.134 0.213 0.211 0.181 0.261
## 16 16 trafford >2.3 m 0.105 0.305 0.190 0.201 0.199and this time there is an effect of gender, but it is smallish, as befits an effect that is not significant.45
\(\blacksquare\)
- What do you think is the most important way in which the lakes differ? (Hint: look at where the biggest predicted probabilities are.)
Solution
Here are the predictions again:
preds1## # A tibble: 8 × 7
## Size Lake Bird Fish Invertebrate Other Reptile
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 <2.3 george 0.0297 0.452 0.413 0.0938 0.0116
## 2 <2.3 hancock 0.0704 0.535 0.0931 0.254 0.0475
## 3 <2.3 oklawaha 0.00882 0.258 0.602 0.0539 0.0772
## 4 <2.3 trafford 0.0359 0.184 0.517 0.174 0.0888
## 5 >2.3 george 0.0811 0.657 0.140 0.0979 0.0239
## 6 >2.3 hancock 0.141 0.570 0.0231 0.194 0.0718
## 7 >2.3 oklawaha 0.0294 0.458 0.249 0.0687 0.195
## 8 >2.3 trafford 0.108 0.296 0.193 0.201 0.202Following my own hint: the preferred diet in George and Hancock lakes is fish, but the preferred diet in Oklawaha and Trafford lakes is (at least sometimes) invertebrates. That is to say, the preferred diet in those last two lakes is less likely to be invertebrates than it is in the first two (comparing for alligators of the same size). This is true for both large and small alligators, as it should be, since there is no interaction in the model. That will do, though you can also note that reptiles are more commonly found in the last two lakes, and birds sometimes appear in the diet in Hancock and Trafford but rarely in the other two lakes.
Another way to think about this is to hold size constant and compare lakes (and then check that it applies to the other size too). In this case, you’d find the biggest predictions among the first four rows, and then check that the pattern persists in the second four rows. (It does.)
I think looking at predicted probabilities like this is the easiest way to see what the model is telling you.
\(\blacksquare\)
- How would you describe the major difference between the diets of the small and large alligators?
Solution
Same idea: hold lake constant, and compare small and large, then check that your conclusion holds for the other lakes as it should. For example, in George Lake, the large alligators are more likely to eat fish, and less likely to eat invertebrates, compared to the small ones. The other food types are not that much different, though you might also note that birds appear more in the diets of large alligators than small ones. Does that hold in the other lakes? I think so, though there is less difference for fish in Hancock lake than the others (where invertebrates are rare for both sizes). Birds don’t commonly appear in any alligator’s diets, but where they do, they are commoner for large alligators than small ones.
\(\blacksquare\)
28.10 Crimes in San Francisco
The data in link is a subset of a huge dataset of crimes committed in San Francisco between 2003 and 2015. The variables are:
Dates: the date and time of the crimeCategory: the category of crime, eg. “larceny” or “vandalism” (response).Descript: detailed description of crime.DayOfWeek: the day of the week of the crime.PdDistrict: the name of the San Francisco Police Department district in which the crime took place.Resolution: how the crime was resolvedAddress: approximate street address of crimeX: longitudeY: latitude
Our aim is to see whether the category of crime depends on the day of the week and the district in which it occurred. However, there are a lot of crime categories, so we will focus on the top four “interesting” ones, which are the ones included in this data file.
Some of the model-fitting takes a while (you’ll see why below). If
you’re using R Markdown, you can wait for the models to fit each time
you re-run your document, or insert cache=T in the top line
of your code chunk (the one with r in curly brackets in it,
above the actual code). Put a comma and the cache=T inside
the curly brackets. What that does is to re-run that code chunk only
if it changes; if it hasn’t changed it will use the saved results from
last time it was run. That can save you a lot of waiting around.
- Read in the data and display the dataset (or, at least, part of it).
Solution
The usual:
my_url <- "http://utsc.utoronto.ca/~butler/d29/sfcrime1.csv"
sfcrime <- read_csv(my_url)## Rows: 359528 Columns: 3
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (3): Category, DayOfWeek, PdDistrict
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.sfcrime## # A tibble: 359,528 × 3
## Category DayOfWeek PdDistrict
## <chr> <chr> <chr>
## 1 LARCENY/THEFT Wednesday NORTHERN
## 2 LARCENY/THEFT Wednesday PARK
## 3 LARCENY/THEFT Wednesday INGLESIDE
## 4 VEHICLE THEFT Wednesday INGLESIDE
## 5 VEHICLE THEFT Wednesday BAYVIEW
## 6 LARCENY/THEFT Wednesday RICHMOND
## 7 LARCENY/THEFT Wednesday CENTRAL
## 8 LARCENY/THEFT Wednesday CENTRAL
## 9 LARCENY/THEFT Wednesday NORTHERN
## 10 ASSAULT Wednesday INGLESIDE
## # … with 359,518 more rowsThis is a tidied-up version of the data, with only the variables we’ll look at, and only the observations from one of the “big four” crimes, a mere 300,000 of them. This is the data set we created earlier.
\(\blacksquare\)
- Fit a multinomial logistic regression that predicts crime category from day of week and district. (You don’t need to look at it.) The model-fitting produces some output. (If you’re using R Markdown, that will come with it.)
Solution
The modelling part is easy enough, as long as you can get the uppercase letters in the right places:
sfcrime.1 <- multinom(Category~DayOfWeek+PdDistrict,data=sfcrime)## # weights: 68 (48 variable)
## initial value 498411.639069
## iter 10 value 430758.073422
## iter 20 value 430314.270403
## iter 30 value 423303.587698
## iter 40 value 420883.528523
## iter 50 value 418355.242764
## final value 418149.979622
## converged\(\blacksquare\)
- Fit a model that predicts Category from only the district. Hand in the output from the fitting process as well.
Solution
Same idea. Write it out, or use update:
sfcrime.2 <- update(sfcrime.1,.~.-DayOfWeek)## # weights: 44 (30 variable)
## initial value 498411.639069
## iter 10 value 426003.543845
## iter 20 value 425542.806828
## iter 30 value 421715.787609
## final value 418858.235297
## converged\(\blacksquare\)
- Use
anovato compare the two models you just obtained. What does theanovatell you?
Solution
This:
anova(sfcrime.2,sfcrime.1)## Likelihood ratio tests of Multinomial Models
##
## Response: Category
## Model Resid. df Resid. Dev Test Df LR stat. Pr(Chi)
## 1 PdDistrict 1078554 837716.5
## 2 DayOfWeek + PdDistrict 1078536 836300.0 1 vs 2 18 1416.511 0This is a very small P-value. The null hypothesis is that the two
models are equally good, and this is clearly rejected. We need the
bigger model: that is, we need to keep DayOfWeek in there,
because the pattern of crimes (in each district) differs over day of
week.
One reason the P-value came out so small is that we have a ton of data, so that even a very small difference between days of the week could come out very strongly significant. The Machine Learning people (this is a machine learning dataset) don’t worry so much about tests for that reason: they are more concerned with predicting things well, so they just throw everything into the model and see what comes out.
\(\blacksquare\)
- Using your preferred model, obtain predicted probabilities
that a crime will be of each of these four categories for each day of
the week in the
TENDERLOINdistrict (the name is ALL CAPS). This will mean constructing a data frame to predict from, obtaining the predictions and then displaying them suitably.
Solution
There is an easier way, which takes a bit of adapting (and takes a bit of time to run):
p <- predictions(sfcrime.1, variables = c("DayOfWeek", "PdDistrict"), type = "probs")
p## rowid type group predicted std.error Category DayOfWeek
## 1 1 probs ASSAULT 0.16740047 0.004249022 LARCENY/THEFT Wednesday
## 2 2 probs ASSAULT 0.17495015 0.007700747 LARCENY/THEFT Wednesday
## 3 3 probs ASSAULT 0.27563912 0.004316150 LARCENY/THEFT Wednesday
## 4 4 probs ASSAULT 0.29800440 0.004808902 LARCENY/THEFT Wednesday
## 5 5 probs ASSAULT 0.17099635 0.007372097 LARCENY/THEFT Wednesday
## 6 6 probs ASSAULT 0.17783225 0.004947000 LARCENY/THEFT Wednesday
## 7 7 probs ASSAULT 0.21299344 0.005677917 LARCENY/THEFT Wednesday
## 8 8 probs ASSAULT 0.17038113 0.006150633 LARCENY/THEFT Wednesday
## 9 9 probs ASSAULT 0.18748667 0.007646783 LARCENY/THEFT Wednesday
## 10 10 probs ASSAULT 0.23214690 0.003291730 LARCENY/THEFT Wednesday
## 11 11 probs ASSAULT 0.16766532 0.002257512 LARCENY/THEFT Tuesday
## 12 12 probs ASSAULT 0.17593330 0.003469069 LARCENY/THEFT Tuesday
## 13 13 probs ASSAULT 0.27614457 0.001866336 LARCENY/THEFT Tuesday
## 14 14 probs ASSAULT 0.30028506 0.001571976 LARCENY/THEFT Tuesday
## 15 15 probs ASSAULT 0.17069994 0.003170870 LARCENY/THEFT Tuesday
## 16 16 probs ASSAULT 0.17733529 0.002258191 LARCENY/THEFT Tuesday
## 17 17 probs ASSAULT 0.21285423 0.002724312 LARCENY/THEFT Tuesday
## 18 18 probs ASSAULT 0.17126617 0.003098840 LARCENY/THEFT Tuesday
## 19 19 probs ASSAULT 0.19424470 0.004026416 LARCENY/THEFT Tuesday
## 20 20 probs ASSAULT 0.23480816 0.001286873 LARCENY/THEFT Tuesday
## 21 21 probs ASSAULT 0.17389840 0.005326866 LARCENY/THEFT Monday
## 22 22 probs ASSAULT 0.18249497 0.008362492 LARCENY/THEFT Monday
## 23 23 probs ASSAULT 0.28336267 0.004541134 LARCENY/THEFT Monday
## 24 24 probs ASSAULT 0.30985102 0.006048981 LARCENY/THEFT Monday
## 25 25 probs ASSAULT 0.17601746 0.008187888 LARCENY/THEFT Monday
## 26 26 probs ASSAULT 0.18337060 0.005174147 LARCENY/THEFT Monday
## 27 27 probs ASSAULT 0.21906616 0.006007794 LARCENY/THEFT Monday
## 28 28 probs ASSAULT 0.17845671 0.006830243 LARCENY/THEFT Monday
## 29 29 probs ASSAULT 0.20724299 0.008414602 LARCENY/THEFT Monday
## 30 30 probs ASSAULT 0.24433533 0.003468537 LARCENY/THEFT Monday
## 31 31 probs ASSAULT 0.19696492 0.003559191 LARCENY/THEFT Sunday
## 32 32 probs ASSAULT 0.20800510 0.005728635 LARCENY/THEFT Sunday
## 33 33 probs ASSAULT 0.31443475 0.003774785 LARCENY/THEFT Sunday
## 34 34 probs ASSAULT 0.34749104 0.003963707 LARCENY/THEFT Sunday
## 35 35 probs ASSAULT 0.19736750 0.005492126 LARCENY/THEFT Sunday
## 36 36 probs ASSAULT 0.20540858 0.004071987 LARCENY/THEFT Sunday
## 37 37 probs ASSAULT 0.24446582 0.005088369 LARCENY/THEFT Sunday
## 38 38 probs ASSAULT 0.20397995 0.005591464 LARCENY/THEFT Sunday
## 39 39 probs ASSAULT 0.25485968 0.007256869 LARCENY/THEFT Sunday
## 40 40 probs ASSAULT 0.27984344 0.002676152 LARCENY/THEFT Sunday
## 41 41 probs ASSAULT 0.18092763 0.004311834 LARCENY/THEFT Saturday
## 42 42 probs ASSAULT 0.19195798 0.006726984 LARCENY/THEFT Saturday
## 43 43 probs ASSAULT 0.29267736 0.005522881 LARCENY/THEFT Saturday
## 44 44 probs ASSAULT 0.32591927 0.004402369 LARCENY/THEFT Saturday
## 45 45 probs ASSAULT 0.18093521 0.006584115 LARCENY/THEFT Saturday
## 46 46 probs ASSAULT 0.18820496 0.005898019 LARCENY/THEFT Saturday
## 47 47 probs ASSAULT 0.22541501 0.006543871 LARCENY/THEFT Saturday
## 48 48 probs ASSAULT 0.18797843 0.007245661 LARCENY/THEFT Saturday
## 49 49 probs ASSAULT 0.24259978 0.008750468 LARCENY/THEFT Saturday
## 50 50 probs ASSAULT 0.26134330 0.003953550 LARCENY/THEFT Saturday
## 51 51 probs ASSAULT 0.16632335 0.003055645 LARCENY/THEFT Friday
## 52 52 probs ASSAULT 0.17517886 0.005516369 LARCENY/THEFT Friday
## 53 53 probs ASSAULT 0.27029845 0.003146449 LARCENY/THEFT Friday
## 54 54 probs ASSAULT 0.29998131 0.002873313 LARCENY/THEFT Friday
## 55 55 probs ASSAULT 0.16663037 0.005263419 LARCENY/THEFT Friday
## 56 56 probs ASSAULT 0.17433914 0.003450540 LARCENY/THEFT Friday
## 57 57 probs ASSAULT 0.20798507 0.004412468 LARCENY/THEFT Friday
## 58 58 probs ASSAULT 0.17233336 0.004688380 LARCENY/THEFT Friday
## 59 59 probs ASSAULT 0.21259967 0.006090589 LARCENY/THEFT Friday
## 60 60 probs ASSAULT 0.23824733 0.002408765 LARCENY/THEFT Friday
## 61 61 probs ASSAULT 0.16586163 0.005495554 LARCENY/THEFT Thursday
## 62 62 probs ASSAULT 0.17387271 0.007175742 LARCENY/THEFT Thursday
## 63 63 probs ASSAULT 0.27237141 0.004860729 LARCENY/THEFT Thursday
## 64 64 probs ASSAULT 0.29706897 0.005153000 LARCENY/THEFT Thursday
## 65 65 probs ASSAULT 0.16838408 0.006982086 LARCENY/THEFT Thursday
## 66 66 probs ASSAULT 0.17539267 0.005469551 LARCENY/THEFT Thursday
## 67 67 probs ASSAULT 0.20998973 0.006113640 LARCENY/THEFT Thursday
## 68 68 probs ASSAULT 0.16979145 0.006498304 LARCENY/THEFT Thursday
## 69 69 probs ASSAULT 0.19387537 0.007413903 LARCENY/THEFT Thursday
## 70 70 probs ASSAULT 0.23270297 0.003658268 LARCENY/THEFT Thursday
## 71 1 probs DRUG/NARCOTIC 0.11430177 0.006284541 LARCENY/THEFT Wednesday
## 72 2 probs DRUG/NARCOTIC 0.16067021 0.003709099 LARCENY/THEFT Wednesday
## 73 3 probs DRUG/NARCOTIC 0.09574189 0.002921544 LARCENY/THEFT Wednesday
## 74 4 probs DRUG/NARCOTIC 0.17019228 0.003543725 LARCENY/THEFT Wednesday
## 75 5 probs DRUG/NARCOTIC 0.06670490 0.003813318 LARCENY/THEFT Wednesday
## 76 6 probs DRUG/NARCOTIC 0.05827926 0.002740684 LARCENY/THEFT Wednesday
## 77 7 probs DRUG/NARCOTIC 0.07436918 0.003443478 LARCENY/THEFT Wednesday
## 78 8 probs DRUG/NARCOTIC 0.16282479 0.003402016 LARCENY/THEFT Wednesday
## 79 9 probs DRUG/NARCOTIC 0.54002868 0.002666268 LARCENY/THEFT Wednesday
## 80 10 probs DRUG/NARCOTIC 0.22943594 0.004915437 LARCENY/THEFT Wednesday
## 81 11 probs DRUG/NARCOTIC 0.10657874 0.008527641 LARCENY/THEFT Tuesday
## 82 12 probs DRUG/NARCOTIC 0.15041812 0.007622952 LARCENY/THEFT Tuesday
## 83 13 probs DRUG/NARCOTIC 0.08929533 0.008338873 LARCENY/THEFT Tuesday
## 84 14 probs DRUG/NARCOTIC 0.15965480 0.007334967 LARCENY/THEFT Tuesday
## 85 15 probs DRUG/NARCOTIC 0.06199196 0.008108777 LARCENY/THEFT Tuesday
## 86 16 probs DRUG/NARCOTIC 0.05410405 0.007388529 LARCENY/THEFT Tuesday
## 87 17 probs DRUG/NARCOTIC 0.06918948 0.008831074 LARCENY/THEFT Tuesday
## 88 18 probs DRUG/NARCOTIC 0.15237078 0.008574787 LARCENY/THEFT Tuesday
## 89 19 probs DRUG/NARCOTIC 0.52086682 0.007446236 LARCENY/THEFT Tuesday
## 90 20 probs DRUG/NARCOTIC 0.21604430 0.010113856 LARCENY/THEFT Tuesday
## 91 21 probs DRUG/NARCOTIC 0.09948621 0.005230659 LARCENY/THEFT Monday
## 92 22 probs DRUG/NARCOTIC 0.14042453 0.002905915 LARCENY/THEFT Monday
## 93 23 probs DRUG/NARCOTIC 0.08246598 0.002204010 LARCENY/THEFT Monday
## 94 24 probs DRUG/NARCOTIC 0.14826586 0.002799410 LARCENY/THEFT Monday
## 95 25 probs DRUG/NARCOTIC 0.05753044 0.002999987 LARCENY/THEFT Monday
## 96 26 probs DRUG/NARCOTIC 0.05035056 0.002009011 LARCENY/THEFT Monday
## 97 27 probs DRUG/NARCOTIC 0.06408746 0.002602336 LARCENY/THEFT Monday
## 98 28 probs DRUG/NARCOTIC 0.14289037 0.002553548 LARCENY/THEFT Monday
## 99 29 probs DRUG/NARCOTIC 0.50014661 0.001959300 LARCENY/THEFT Monday
## 100 30 probs DRUG/NARCOTIC 0.20232795 0.003720120 LARCENY/THEFT Monday
## 101 31 probs DRUG/NARCOTIC 0.07854536 0.008107965 LARCENY/THEFT Sunday
## 102 32 probs DRUG/NARCOTIC 0.11156563 0.005288932 LARCENY/THEFT Sunday
## 103 33 probs DRUG/NARCOTIC 0.06378625 0.004092791 LARCENY/THEFT Sunday
## 104 34 probs DRUG/NARCOTIC 0.11590340 0.005041854 LARCENY/THEFT Sunday
## 105 35 probs DRUG/NARCOTIC 0.04496576 0.005449351 LARCENY/THEFT Sunday
## 106 36 probs DRUG/NARCOTIC 0.03931493 0.003862875 LARCENY/THEFT Sunday
## 107 37 probs DRUG/NARCOTIC 0.04985173 0.004826396 LARCENY/THEFT Sunday
## 108 38 probs DRUG/NARCOTIC 0.11384708 0.004732057 LARCENY/THEFT Sunday
## 109 39 probs DRUG/NARCOTIC 0.42872910 0.003948120 LARCENY/THEFT Sunday
## 110 40 probs DRUG/NARCOTIC 0.16152847 0.006018277 LARCENY/THEFT Sunday
## 111 41 probs DRUG/NARCOTIC 0.07426621 0.008310284 LARCENY/THEFT Saturday
## 112 42 probs DRUG/NARCOTIC 0.10597839 0.005931533 LARCENY/THEFT Saturday
## 113 43 probs DRUG/NARCOTIC 0.06111395 0.005196966 LARCENY/THEFT Saturday
## 114 44 probs DRUG/NARCOTIC 0.11189671 0.005614768 LARCENY/THEFT Saturday
## 115 45 probs DRUG/NARCOTIC 0.04243108 0.006053404 LARCENY/THEFT Saturday
## 116 46 probs DRUG/NARCOTIC 0.03707872 0.005344226 LARCENY/THEFT Saturday
## 117 47 probs DRUG/NARCOTIC 0.04731509 0.005799895 LARCENY/THEFT Saturday
## 118 48 probs DRUG/NARCOTIC 0.10799338 0.005347127 LARCENY/THEFT Saturday
## 119 49 probs DRUG/NARCOTIC 0.42007509 0.004293186 LARCENY/THEFT Saturday
## 120 50 probs DRUG/NARCOTIC 0.15527446 0.008273414 LARCENY/THEFT Saturday
## 121 51 probs DRUG/NARCOTIC 0.08638758 0.006242735 LARCENY/THEFT Friday
## 122 52 probs DRUG/NARCOTIC 0.12237835 0.003771573 LARCENY/THEFT Friday
## 123 53 probs DRUG/NARCOTIC 0.07141782 0.002931476 LARCENY/THEFT Friday
## 124 54 probs DRUG/NARCOTIC 0.13032065 0.003578776 LARCENY/THEFT Friday
## 125 55 probs DRUG/NARCOTIC 0.04944552 0.003915027 LARCENY/THEFT Friday
## 126 56 probs DRUG/NARCOTIC 0.04346106 0.002744315 LARCENY/THEFT Friday
## 127 57 probs DRUG/NARCOTIC 0.05524090 0.003446647 LARCENY/THEFT Friday
## 128 58 probs DRUG/NARCOTIC 0.12527668 0.003379966 LARCENY/THEFT Friday
## 129 59 probs DRUG/NARCOTIC 0.46581221 0.002597267 LARCENY/THEFT Friday
## 130 60 probs DRUG/NARCOTIC 0.17911356 0.004828833 LARCENY/THEFT Friday
## 131 61 probs DRUG/NARCOTIC 0.10504150 0.006327675 LARCENY/THEFT Thursday
## 132 62 probs DRUG/NARCOTIC 0.14810551 0.003638651 LARCENY/THEFT Thursday
## 133 63 probs DRUG/NARCOTIC 0.08774885 0.002902641 LARCENY/THEFT Thursday
## 134 64 probs DRUG/NARCOTIC 0.15735959 0.003600833 LARCENY/THEFT Thursday
## 135 65 probs DRUG/NARCOTIC 0.06092432 0.003776777 LARCENY/THEFT Thursday
## 136 66 probs DRUG/NARCOTIC 0.05331308 0.002908602 LARCENY/THEFT Thursday
## 137 67 probs DRUG/NARCOTIC 0.06800542 0.003052086 LARCENY/THEFT Thursday
## 138 68 probs DRUG/NARCOTIC 0.15049900 0.002863440 LARCENY/THEFT Thursday
## 139 69 probs DRUG/NARCOTIC 0.51795000 0.001949593 LARCENY/THEFT Thursday
## 140 70 probs DRUG/NARCOTIC 0.21331394 0.004079474 LARCENY/THEFT Thursday
## 141 1 probs LARCENY/THEFT 0.59175834 0.007984061 LARCENY/THEFT Wednesday
## 142 2 probs LARCENY/THEFT 0.46733060 0.006390188 LARCENY/THEFT Wednesday
## 143 3 probs LARCENY/THEFT 0.33934630 0.005653598 LARCENY/THEFT Wednesday
## 144 4 probs LARCENY/THEFT 0.31385481 0.005484272 LARCENY/THEFT Wednesday
## 145 5 probs LARCENY/THEFT 0.54243172 0.006886299 LARCENY/THEFT Wednesday
## 146 6 probs LARCENY/THEFT 0.65657190 0.004909500 LARCENY/THEFT Wednesday
## 147 7 probs LARCENY/THEFT 0.47347628 0.008217094 LARCENY/THEFT Wednesday
## 148 8 probs LARCENY/THEFT 0.60079333 0.007680014 LARCENY/THEFT Wednesday
## 149 9 probs LARCENY/THEFT 0.24794975 0.009270139 LARCENY/THEFT Wednesday
## 150 10 probs LARCENY/THEFT 0.38966273 0.007904725 LARCENY/THEFT Wednesday
## 151 11 probs LARCENY/THEFT 0.59833785 0.005173013 LARCENY/THEFT Tuesday
## 152 12 probs LARCENY/THEFT 0.47443144 0.005660640 LARCENY/THEFT Tuesday
## 153 13 probs LARCENY/THEFT 0.34320556 0.005313942 LARCENY/THEFT Tuesday
## 154 14 probs LARCENY/THEFT 0.31926799 0.005081944 LARCENY/THEFT Tuesday
## 155 15 probs LARCENY/THEFT 0.54664723 0.005920510 LARCENY/THEFT Tuesday
## 156 16 probs LARCENY/THEFT 0.66097109 0.004468250 LARCENY/THEFT Tuesday
## 157 17 probs LARCENY/THEFT 0.47767204 0.007550911 LARCENY/THEFT Tuesday
## 158 18 probs LARCENY/THEFT 0.60966429 0.007305707 LARCENY/THEFT Tuesday
## 159 19 probs LARCENY/THEFT 0.25933313 0.008657594 LARCENY/THEFT Tuesday
## 160 20 probs LARCENY/THEFT 0.39788238 0.007471666 LARCENY/THEFT Tuesday
## 161 21 probs LARCENY/THEFT 0.59573012 0.009559346 LARCENY/THEFT Monday
## 162 22 probs LARCENY/THEFT 0.47241866 0.008177660 LARCENY/THEFT Monday
## 163 23 probs LARCENY/THEFT 0.33807352 0.006940633 LARCENY/THEFT Monday
## 164 24 probs LARCENY/THEFT 0.31624620 0.006927657 LARCENY/THEFT Monday
## 165 25 probs LARCENY/THEFT 0.54110340 0.008026769 LARCENY/THEFT Monday
## 166 26 probs LARCENY/THEFT 0.65609650 0.006192411 LARCENY/THEFT Monday
## 167 27 probs LARCENY/THEFT 0.47192561 0.009510114 LARCENY/THEFT Monday
## 168 28 probs LARCENY/THEFT 0.60982156 0.008758318 LARCENY/THEFT Monday
## 169 29 probs LARCENY/THEFT 0.26560692 0.009942019 LARCENY/THEFT Monday
## 170 30 probs LARCENY/THEFT 0.39744635 0.008844026 LARCENY/THEFT Monday
## 171 31 probs LARCENY/THEFT 0.59260407 0.009033232 LARCENY/THEFT Sunday
## 172 32 probs LARCENY/THEFT 0.47290283 0.007531301 LARCENY/THEFT Sunday
## 173 33 probs LARCENY/THEFT 0.32947377 0.006474767 LARCENY/THEFT Sunday
## 174 34 probs LARCENY/THEFT 0.31148545 0.006792907 LARCENY/THEFT Sunday
## 175 35 probs LARCENY/THEFT 0.53287082 0.007784299 LARCENY/THEFT Sunday
## 176 36 probs LARCENY/THEFT 0.64547345 0.005651106 LARCENY/THEFT Sunday
## 177 37 probs LARCENY/THEFT 0.46252816 0.008448728 LARCENY/THEFT Sunday
## 178 38 probs LARCENY/THEFT 0.61218000 0.009038393 LARCENY/THEFT Sunday
## 179 39 probs LARCENY/THEFT 0.28686823 0.010263234 LARCENY/THEFT Sunday
## 180 40 probs LARCENY/THEFT 0.39978745 0.009370157 LARCENY/THEFT Sunday
## 181 41 probs LARCENY/THEFT 0.61012247 0.008609439 LARCENY/THEFT Saturday
## 182 42 probs LARCENY/THEFT 0.48914822 0.007464901 LARCENY/THEFT Saturday
## 183 43 probs LARCENY/THEFT 0.34372871 0.007305465 LARCENY/THEFT Saturday
## 184 44 probs LARCENY/THEFT 0.32744667 0.006962282 LARCENY/THEFT Saturday
## 185 45 probs LARCENY/THEFT 0.54752727 0.007613625 LARCENY/THEFT Saturday
## 186 46 probs LARCENY/THEFT 0.66286830 0.005204388 LARCENY/THEFT Saturday
## 187 47 probs LARCENY/THEFT 0.47801249 0.009458660 LARCENY/THEFT Saturday
## 188 48 probs LARCENY/THEFT 0.63231874 0.009280756 LARCENY/THEFT Saturday
## 189 49 probs LARCENY/THEFT 0.30606108 0.011494559 LARCENY/THEFT Saturday
## 190 50 probs LARCENY/THEFT 0.41846755 0.009767265 LARCENY/THEFT Saturday
## 191 51 probs LARCENY/THEFT 0.60806299 0.007546729 LARCENY/THEFT Friday
## 192 52 probs LARCENY/THEFT 0.48394860 0.005984849 LARCENY/THEFT Friday
## 193 53 probs LARCENY/THEFT 0.34415452 0.005240767 LARCENY/THEFT Friday
## 194 54 probs LARCENY/THEFT 0.32674427 0.004696771 LARCENY/THEFT Friday
## 195 55 probs LARCENY/THEFT 0.54666349 0.006314808 LARCENY/THEFT Friday
## 196 56 probs LARCENY/THEFT 0.66569350 0.004751163 LARCENY/THEFT Friday
## 197 57 probs LARCENY/THEFT 0.47815843 0.007875598 LARCENY/THEFT Friday
## 198 58 probs LARCENY/THEFT 0.62846433 0.006871688 LARCENY/THEFT Friday
## 199 59 probs LARCENY/THEFT 0.29077933 0.007962724 LARCENY/THEFT Friday
## 200 60 probs LARCENY/THEFT 0.41358206 0.007362817 LARCENY/THEFT Friday
## 201 61 probs LARCENY/THEFT 0.59859147 0.010281887 LARCENY/THEFT Thursday
## 202 62 probs LARCENY/THEFT 0.47417453 0.008878386 LARCENY/THEFT Thursday
## 203 63 probs LARCENY/THEFT 0.34234239 0.008578470 LARCENY/THEFT Thursday
## 204 64 probs LARCENY/THEFT 0.31941867 0.007577982 LARCENY/THEFT Thursday
## 205 65 probs LARCENY/THEFT 0.54532596 0.009163512 LARCENY/THEFT Thursday
## 206 66 probs LARCENY/THEFT 0.66111973 0.008568597 LARCENY/THEFT Thursday
## 207 67 probs LARCENY/THEFT 0.47657026 0.009999979 LARCENY/THEFT Thursday
## 208 68 probs LARCENY/THEFT 0.61124644 0.008665315 LARCENY/THEFT Thursday
## 209 69 probs LARCENY/THEFT 0.26176575 0.008555238 LARCENY/THEFT Thursday
## 210 70 probs LARCENY/THEFT 0.39877214 0.008349837 LARCENY/THEFT Thursday
## 211 1 probs VEHICLE THEFT 0.12653942 0.004778899 LARCENY/THEFT Wednesday
## 212 2 probs VEHICLE THEFT 0.19704904 0.005426455 LARCENY/THEFT Wednesday
## 213 3 probs VEHICLE THEFT 0.28927268 0.007319912 LARCENY/THEFT Wednesday
## 214 4 probs VEHICLE THEFT 0.21794851 0.004875745 LARCENY/THEFT Wednesday
## 215 5 probs VEHICLE THEFT 0.21986703 0.005971657 LARCENY/THEFT Wednesday
## 216 6 probs VEHICLE THEFT 0.10731659 0.007864579 LARCENY/THEFT Wednesday
## 217 7 probs VEHICLE THEFT 0.23916110 0.008140716 LARCENY/THEFT Wednesday
## 218 8 probs VEHICLE THEFT 0.06600076 0.005079759 LARCENY/THEFT Wednesday
## 219 9 probs VEHICLE THEFT 0.02453490 0.002094417 LARCENY/THEFT Wednesday
## 220 10 probs VEHICLE THEFT 0.14875443 0.006893103 LARCENY/THEFT Wednesday
## 221 11 probs VEHICLE THEFT 0.12741809 0.003022617 LARCENY/THEFT Tuesday
## 222 12 probs VEHICLE THEFT 0.19921715 0.004393776 LARCENY/THEFT Tuesday
## 223 13 probs VEHICLE THEFT 0.29135453 0.008232705 LARCENY/THEFT Tuesday
## 224 14 probs VEHICLE THEFT 0.22079215 0.004383307 LARCENY/THEFT Tuesday
## 225 15 probs VEHICLE THEFT 0.22066087 0.004907536 LARCENY/THEFT Tuesday
## 226 16 probs VEHICLE THEFT 0.10758957 0.008930829 LARCENY/THEFT Tuesday
## 227 17 probs VEHICLE THEFT 0.24028425 0.007038654 LARCENY/THEFT Tuesday
## 228 18 probs VEHICLE THEFT 0.06669875 0.004251317 LARCENY/THEFT Tuesday
## 229 19 probs VEHICLE THEFT 0.02555535 0.001469659 LARCENY/THEFT Tuesday
## 230 20 probs VEHICLE THEFT 0.15126516 0.006274676 LARCENY/THEFT Tuesday
## 231 21 probs VEHICLE THEFT 0.13088527 0.005619880 LARCENY/THEFT Monday
## 232 22 probs VEHICLE THEFT 0.20466185 0.006133126 LARCENY/THEFT Monday
## 233 23 probs VEHICLE THEFT 0.29609782 0.008063400 LARCENY/THEFT Monday
## 234 24 probs VEHICLE THEFT 0.22563692 0.005675618 LARCENY/THEFT Monday
## 235 25 probs VEHICLE THEFT 0.22534869 0.006536327 LARCENY/THEFT Monday
## 236 26 probs VEHICLE THEFT 0.11018235 0.008545907 LARCENY/THEFT Monday
## 237 27 probs VEHICLE THEFT 0.24492077 0.008849760 LARCENY/THEFT Monday
## 238 28 probs VEHICLE THEFT 0.06883136 0.005615097 LARCENY/THEFT Monday
## 239 29 probs VEHICLE THEFT 0.02700348 0.002201869 LARCENY/THEFT Monday
## 240 30 probs VEHICLE THEFT 0.15589038 0.007687735 LARCENY/THEFT Monday
## 241 31 probs VEHICLE THEFT 0.13188565 0.006641383 LARCENY/THEFT Sunday
## 242 32 probs VEHICLE THEFT 0.20752645 0.007785445 LARCENY/THEFT Sunday
## 243 33 probs VEHICLE THEFT 0.29230523 0.009429464 LARCENY/THEFT Sunday
## 244 34 probs VEHICLE THEFT 0.22512011 0.007509519 LARCENY/THEFT Sunday
## 245 35 probs VEHICLE THEFT 0.22479591 0.008327058 LARCENY/THEFT Sunday
## 246 36 probs VEHICLE THEFT 0.10980304 0.010221023 LARCENY/THEFT Sunday
## 247 37 probs VEHICLE THEFT 0.24315429 0.009800940 LARCENY/THEFT Sunday
## 248 38 probs VEHICLE THEFT 0.06999296 0.007301930 LARCENY/THEFT Sunday
## 249 39 probs VEHICLE THEFT 0.02954299 0.002952967 LARCENY/THEFT Sunday
## 250 40 probs VEHICLE THEFT 0.15884064 0.008979404 LARCENY/THEFT Sunday
## 251 41 probs VEHICLE THEFT 0.13468369 0.004122828 LARCENY/THEFT Saturday
## 252 42 probs VEHICLE THEFT 0.21291541 0.005337961 LARCENY/THEFT Saturday
## 253 43 probs VEHICLE THEFT 0.30247998 0.009106063 LARCENY/THEFT Saturday
## 254 44 probs VEHICLE THEFT 0.23473735 0.005329537 LARCENY/THEFT Saturday
## 255 45 probs VEHICLE THEFT 0.22910644 0.005736426 LARCENY/THEFT Saturday
## 256 46 probs VEHICLE THEFT 0.11184803 0.010481614 LARCENY/THEFT Saturday
## 257 47 probs VEHICLE THEFT 0.24925741 0.008383304 LARCENY/THEFT Saturday
## 258 48 probs VEHICLE THEFT 0.07170945 0.005065664 LARCENY/THEFT Saturday
## 259 49 probs VEHICLE THEFT 0.03126405 0.001761442 LARCENY/THEFT Saturday
## 260 50 probs VEHICLE THEFT 0.16491469 0.007913243 LARCENY/THEFT Saturday
## 261 51 probs VEHICLE THEFT 0.13922608 0.004256068 LARCENY/THEFT Friday
## 262 52 probs VEHICLE THEFT 0.21849420 0.004514973 LARCENY/THEFT Friday
## 263 53 probs VEHICLE THEFT 0.31412922 0.006302301 LARCENY/THEFT Friday
## 264 54 probs VEHICLE THEFT 0.24295377 0.003783108 LARCENY/THEFT Friday
## 265 55 probs VEHICLE THEFT 0.23726062 0.005015415 LARCENY/THEFT Friday
## 266 56 probs VEHICLE THEFT 0.11650631 0.007255296 LARCENY/THEFT Friday
## 267 57 probs VEHICLE THEFT 0.25861559 0.007256625 LARCENY/THEFT Friday
## 268 58 probs VEHICLE THEFT 0.07392563 0.004209260 LARCENY/THEFT Friday
## 269 59 probs VEHICLE THEFT 0.03080879 0.001798800 LARCENY/THEFT Friday
## 270 60 probs VEHICLE THEFT 0.16905705 0.005939092 LARCENY/THEFT Friday
## 271 61 probs VEHICLE THEFT 0.13050540 0.003565843 LARCENY/THEFT Thursday
## 272 62 probs VEHICLE THEFT 0.20384724 0.004193237 LARCENY/THEFT Thursday
## 273 63 probs VEHICLE THEFT 0.29753735 0.008036866 LARCENY/THEFT Thursday
## 274 64 probs VEHICLE THEFT 0.22615277 0.004283926 LARCENY/THEFT Thursday
## 275 65 probs VEHICLE THEFT 0.22536564 0.004681631 LARCENY/THEFT Thursday
## 276 66 probs VEHICLE THEFT 0.11017452 0.009945436 LARCENY/THEFT Thursday
## 277 67 probs VEHICLE THEFT 0.24543459 0.006572338 LARCENY/THEFT Thursday
## 278 68 probs VEHICLE THEFT 0.06846312 0.003690960 LARCENY/THEFT Thursday
## 279 69 probs VEHICLE THEFT 0.02640888 0.001026810 LARCENY/THEFT Thursday
## 280 70 probs VEHICLE THEFT 0.15521095 0.006099243 LARCENY/THEFT Thursday
## PdDistrict
## 1 NORTHERN
## 2 PARK
## 3 INGLESIDE
## 4 BAYVIEW
## 5 RICHMOND
## 6 CENTRAL
## 7 TARAVAL
## 8 SOUTHERN
## 9 TENDERLOIN
## 10 MISSION
## 11 NORTHERN
## 12 PARK
## 13 INGLESIDE
## 14 BAYVIEW
## 15 RICHMOND
## 16 CENTRAL
## 17 TARAVAL
## 18 SOUTHERN
## 19 TENDERLOIN
## 20 MISSION
## 21 NORTHERN
## 22 PARK
## 23 INGLESIDE
## 24 BAYVIEW
## 25 RICHMOND
## 26 CENTRAL
## 27 TARAVAL
## 28 SOUTHERN
## 29 TENDERLOIN
## 30 MISSION
## 31 NORTHERN
## 32 PARK
## 33 INGLESIDE
## 34 BAYVIEW
## 35 RICHMOND
## 36 CENTRAL
## 37 TARAVAL
## 38 SOUTHERN
## 39 TENDERLOIN
## 40 MISSION
## 41 NORTHERN
## 42 PARK
## 43 INGLESIDE
## 44 BAYVIEW
## 45 RICHMOND
## 46 CENTRAL
## 47 TARAVAL
## 48 SOUTHERN
## 49 TENDERLOIN
## 50 MISSION
## 51 NORTHERN
## 52 PARK
## 53 INGLESIDE
## 54 BAYVIEW
## 55 RICHMOND
## 56 CENTRAL
## 57 TARAVAL
## 58 SOUTHERN
## 59 TENDERLOIN
## 60 MISSION
## 61 NORTHERN
## 62 PARK
## 63 INGLESIDE
## 64 BAYVIEW
## 65 RICHMOND
## 66 CENTRAL
## 67 TARAVAL
## 68 SOUTHERN
## 69 TENDERLOIN
## 70 MISSION
## 71 NORTHERN
## 72 PARK
## 73 INGLESIDE
## 74 BAYVIEW
## 75 RICHMOND
## 76 CENTRAL
## 77 TARAVAL
## 78 SOUTHERN
## 79 TENDERLOIN
## 80 MISSION
## 81 NORTHERN
## 82 PARK
## 83 INGLESIDE
## 84 BAYVIEW
## 85 RICHMOND
## 86 CENTRAL
## 87 TARAVAL
## 88 SOUTHERN
## 89 TENDERLOIN
## 90 MISSION
## 91 NORTHERN
## 92 PARK
## 93 INGLESIDE
## 94 BAYVIEW
## 95 RICHMOND
## 96 CENTRAL
## 97 TARAVAL
## 98 SOUTHERN
## 99 TENDERLOIN
## 100 MISSION
## 101 NORTHERN
## 102 PARK
## 103 INGLESIDE
## 104 BAYVIEW
## 105 RICHMOND
## 106 CENTRAL
## 107 TARAVAL
## 108 SOUTHERN
## 109 TENDERLOIN
## 110 MISSION
## 111 NORTHERN
## 112 PARK
## 113 INGLESIDE
## 114 BAYVIEW
## 115 RICHMOND
## 116 CENTRAL
## 117 TARAVAL
## 118 SOUTHERN
## 119 TENDERLOIN
## 120 MISSION
## 121 NORTHERN
## 122 PARK
## 123 INGLESIDE
## 124 BAYVIEW
## 125 RICHMOND
## 126 CENTRAL
## 127 TARAVAL
## 128 SOUTHERN
## 129 TENDERLOIN
## 130 MISSION
## 131 NORTHERN
## 132 PARK
## 133 INGLESIDE
## 134 BAYVIEW
## 135 RICHMOND
## 136 CENTRAL
## 137 TARAVAL
## 138 SOUTHERN
## 139 TENDERLOIN
## 140 MISSION
## 141 NORTHERN
## 142 PARK
## 143 INGLESIDE
## 144 BAYVIEW
## 145 RICHMOND
## 146 CENTRAL
## 147 TARAVAL
## 148 SOUTHERN
## 149 TENDERLOIN
## 150 MISSION
## 151 NORTHERN
## 152 PARK
## 153 INGLESIDE
## 154 BAYVIEW
## 155 RICHMOND
## 156 CENTRAL
## 157 TARAVAL
## 158 SOUTHERN
## 159 TENDERLOIN
## 160 MISSION
## 161 NORTHERN
## 162 PARK
## 163 INGLESIDE
## 164 BAYVIEW
## 165 RICHMOND
## 166 CENTRAL
## 167 TARAVAL
## 168 SOUTHERN
## 169 TENDERLOIN
## 170 MISSION
## 171 NORTHERN
## 172 PARK
## 173 INGLESIDE
## 174 BAYVIEW
## 175 RICHMOND
## 176 CENTRAL
## 177 TARAVAL
## 178 SOUTHERN
## 179 TENDERLOIN
## 180 MISSION
## 181 NORTHERN
## 182 PARK
## 183 INGLESIDE
## 184 BAYVIEW
## 185 RICHMOND
## 186 CENTRAL
## 187 TARAVAL
## 188 SOUTHERN
## 189 TENDERLOIN
## 190 MISSION
## 191 NORTHERN
## 192 PARK
## 193 INGLESIDE
## 194 BAYVIEW
## 195 RICHMOND
## 196 CENTRAL
## 197 TARAVAL
## 198 SOUTHERN
## 199 TENDERLOIN
## 200 MISSION
## 201 NORTHERN
## 202 PARK
## 203 INGLESIDE
## 204 BAYVIEW
## 205 RICHMOND
## 206 CENTRAL
## 207 TARAVAL
## 208 SOUTHERN
## 209 TENDERLOIN
## 210 MISSION
## 211 NORTHERN
## 212 PARK
## 213 INGLESIDE
## 214 BAYVIEW
## 215 RICHMOND
## 216 CENTRAL
## 217 TARAVAL
## 218 SOUTHERN
## 219 TENDERLOIN
## 220 MISSION
## 221 NORTHERN
## 222 PARK
## 223 INGLESIDE
## 224 BAYVIEW
## 225 RICHMOND
## 226 CENTRAL
## 227 TARAVAL
## 228 SOUTHERN
## 229 TENDERLOIN
## 230 MISSION
## 231 NORTHERN
## 232 PARK
## 233 INGLESIDE
## 234 BAYVIEW
## 235 RICHMOND
## 236 CENTRAL
## 237 TARAVAL
## 238 SOUTHERN
## 239 TENDERLOIN
## 240 MISSION
## 241 NORTHERN
## 242 PARK
## 243 INGLESIDE
## 244 BAYVIEW
## 245 RICHMOND
## 246 CENTRAL
## 247 TARAVAL
## 248 SOUTHERN
## 249 TENDERLOIN
## 250 MISSION
## 251 NORTHERN
## 252 PARK
## 253 INGLESIDE
## 254 BAYVIEW
## 255 RICHMOND
## 256 CENTRAL
## 257 TARAVAL
## 258 SOUTHERN
## 259 TENDERLOIN
## 260 MISSION
## 261 NORTHERN
## 262 PARK
## 263 INGLESIDE
## 264 BAYVIEW
## 265 RICHMOND
## 266 CENTRAL
## 267 TARAVAL
## 268 SOUTHERN
## 269 TENDERLOIN
## 270 MISSION
## 271 NORTHERN
## 272 PARK
## 273 INGLESIDE
## 274 BAYVIEW
## 275 RICHMOND
## 276 CENTRAL
## 277 TARAVAL
## 278 SOUTHERN
## 279 TENDERLOIN
## 280 MISSIONThis is because it includes all the districts, not just the one we want, so:
- grab only the district we want
- (as usual for these) grab only the columns we want
- pivot wider to get a column for each group (crime) we want a predicted probability for:
p %>% filter(PdDistrict == "TENDERLOIN") %>%
select(group, predicted, DayOfWeek) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 7 × 5
## DayOfWeek ASSAULT `DRUG/NARCOTIC` `LARCENY/THEFT` `VEHICLE THEFT`
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Wednesday 0.187 0.540 0.248 0.0245
## 2 Tuesday 0.194 0.521 0.259 0.0256
## 3 Monday 0.207 0.500 0.266 0.0270
## 4 Sunday 0.255 0.429 0.287 0.0295
## 5 Saturday 0.243 0.420 0.306 0.0313
## 6 Friday 0.213 0.466 0.291 0.0308
## 7 Thursday 0.194 0.518 0.262 0.0264A better way to do this is to use datagrid first to get the combinations you want (and only those), namely all the days of the week, but only the district called TENDERLOIN.46
So, let’s get the days of the week. The easiest way is to count them and ignore the counts:47
sfcrime %>% count(DayOfWeek) %>%
pull(DayOfWeek) -> daysofweek
daysofweek## [1] "Friday" "Monday" "Saturday" "Sunday" "Thursday" "Tuesday"
## [7] "Wednesday"Now we can use these in datagrid:48
new <- datagrid(model = sfcrime.1, DayOfWeek = daysofweek, PdDistrict = "TENDERLOIN")
new## DayOfWeek PdDistrict
## 1: Friday TENDERLOIN
## 2: Monday TENDERLOIN
## 3: Saturday TENDERLOIN
## 4: Sunday TENDERLOIN
## 5: Thursday TENDERLOIN
## 6: Tuesday TENDERLOIN
## 7: Wednesday TENDERLOINGood. And then predict for just these. This is slow, but not as slow as before. I’m saving the result of this slow part, so that it doesn’t matter if I change my mind later about what to do with it. I want to make sure that I don’t have to do it again, is all:49
p <- predictions(sfcrime.1, newdata = new, type = "probs")
p## rowid type group predicted std.error DayOfWeek PdDistrict
## 1 1 probs ASSAULT 0.21259967 0.006090589 Friday TENDERLOIN
## 2 2 probs ASSAULT 0.20724299 0.008414602 Monday TENDERLOIN
## 3 3 probs ASSAULT 0.24259978 0.008750468 Saturday TENDERLOIN
## 4 4 probs ASSAULT 0.25485968 0.007256869 Sunday TENDERLOIN
## 5 5 probs ASSAULT 0.19387537 0.007413903 Thursday TENDERLOIN
## 6 6 probs ASSAULT 0.19424470 0.004026416 Tuesday TENDERLOIN
## 7 7 probs ASSAULT 0.18748667 0.007646783 Wednesday TENDERLOIN
## 8 1 probs DRUG/NARCOTIC 0.46581221 0.002597267 Friday TENDERLOIN
## 9 2 probs DRUG/NARCOTIC 0.50014661 0.001959300 Monday TENDERLOIN
## 10 3 probs DRUG/NARCOTIC 0.42007509 0.004293186 Saturday TENDERLOIN
## 11 4 probs DRUG/NARCOTIC 0.42872910 0.003948120 Sunday TENDERLOIN
## 12 5 probs DRUG/NARCOTIC 0.51795000 0.001949593 Thursday TENDERLOIN
## 13 6 probs DRUG/NARCOTIC 0.52086682 0.007446236 Tuesday TENDERLOIN
## 14 7 probs DRUG/NARCOTIC 0.54002868 0.002666268 Wednesday TENDERLOIN
## 15 1 probs LARCENY/THEFT 0.29077933 0.007962724 Friday TENDERLOIN
## 16 2 probs LARCENY/THEFT 0.26560692 0.009942019 Monday TENDERLOIN
## 17 3 probs LARCENY/THEFT 0.30606108 0.011494559 Saturday TENDERLOIN
## 18 4 probs LARCENY/THEFT 0.28686823 0.010263234 Sunday TENDERLOIN
## 19 5 probs LARCENY/THEFT 0.26176575 0.008555238 Thursday TENDERLOIN
## 20 6 probs LARCENY/THEFT 0.25933313 0.008657594 Tuesday TENDERLOIN
## 21 7 probs LARCENY/THEFT 0.24794975 0.009270139 Wednesday TENDERLOIN
## 22 1 probs VEHICLE THEFT 0.03080879 0.001798800 Friday TENDERLOIN
## 23 2 probs VEHICLE THEFT 0.02700348 0.002201869 Monday TENDERLOIN
## 24 3 probs VEHICLE THEFT 0.03126405 0.001761442 Saturday TENDERLOIN
## 25 4 probs VEHICLE THEFT 0.02954299 0.002952967 Sunday TENDERLOIN
## 26 5 probs VEHICLE THEFT 0.02640888 0.001026810 Thursday TENDERLOIN
## 27 6 probs VEHICLE THEFT 0.02555535 0.001469659 Tuesday TENDERLOIN
## 28 7 probs VEHICLE THEFT 0.02453490 0.002094417 Wednesday TENDERLOINThis, as you remember, is long format, so grab the columns you need from it and pivot wider. The easiest grab is to just get rid of the standard error column (since you want only one number to pivot-wider):
p %>%
select(-std.error) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 7 × 8
## rowid type DayOfWeek PdDistrict ASSAULT `DRUG/NARCOTIC` `LARCENY/THEFT`
## <int> <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 1 probs Friday TENDERLOIN 0.213 0.466 0.291
## 2 2 probs Monday TENDERLOIN 0.207 0.500 0.266
## 3 3 probs Saturday TENDERLOIN 0.243 0.420 0.306
## 4 4 probs Sunday TENDERLOIN 0.255 0.429 0.287
## 5 5 probs Thursday TENDERLOIN 0.194 0.518 0.262
## 6 6 probs Tuesday TENDERLOIN 0.194 0.521 0.259
## 7 7 probs Wednesday TENDERLOIN 0.187 0.540 0.248
## # … with 1 more variable: `VEHICLE THEFT` <dbl>Success. (If you don’t get rid of the standard errors, you still have 28 rows and a bunch of missing values; the standard errors are unique, and so pivot_wider will infer that everything should be in its own row.)
\(\blacksquare\)
- Describe briefly how the weekend days Saturday and Sunday differ from the rest.
Solution
The days ended up in some quasi-random order, but Saturday and Sunday are still together, so we can still easily compare them with the rest. My take is that the last two columns don’t differ much between weekday and weekend, but the first two columns do: the probability of a crime being an assault is a bit higher on the weekend, and the probability of a crime being drug-related is a bit lower. I will accept anything reasonable supported by the predictions you got. We said there was a strongly significant day-of-week effect, but the changes from weekday to weekend are actually pretty small (but the changes from one weekday to another are even smaller). This supports what I guessed before, that with this much data even a small effect (the one shown here) is statistically significant.50
Extra: I want to compare another district. What districts do we have?
sfcrime %>% count(PdDistrict)## # A tibble: 10 × 2
## PdDistrict n
## <chr> <int>
## 1 BAYVIEW 31693
## 2 CENTRAL 38052
## 3 INGLESIDE 30102
## 4 MISSION 45277
## 5 NORTHERN 47750
## 6 PARK 19197
## 7 RICHMOND 18211
## 8 SOUTHERN 67981
## 9 TARAVAL 24981
## 10 TENDERLOIN 36284This is the number of our “big four” crimes committed in each
district. Let’s look at the lowest-crime district RICHMOND. I
copy and paste my code. Since I want to compare two districts, I
include both:
new <- datagrid(model = sfcrime.1, PdDistrict = c("TENDERLOIN", "RICHMOND"), DayOfWeek = daysofweek)
new## PdDistrict DayOfWeek
## 1: TENDERLOIN Friday
## 2: TENDERLOIN Monday
## 3: TENDERLOIN Saturday
## 4: TENDERLOIN Sunday
## 5: TENDERLOIN Thursday
## 6: TENDERLOIN Tuesday
## 7: TENDERLOIN Wednesday
## 8: RICHMOND Friday
## 9: RICHMOND Monday
## 10: RICHMOND Saturday
## 11: RICHMOND Sunday
## 12: RICHMOND Thursday
## 13: RICHMOND Tuesday
## 14: RICHMOND Wednesdayand then as we just did. I’m going to be a bit more selective about the columns I keep this time, since the display will be a bit wider and I don’t want it to be too big for the page:
p <- predictions(sfcrime.1, newdata = new, type = "probs")
p## rowid type group predicted std.error PdDistrict DayOfWeek
## 1 1 probs ASSAULT 0.21259967 0.006090589 TENDERLOIN Friday
## 2 2 probs ASSAULT 0.20724299 0.008414602 TENDERLOIN Monday
## 3 3 probs ASSAULT 0.24259978 0.008750468 TENDERLOIN Saturday
## 4 4 probs ASSAULT 0.25485968 0.007256869 TENDERLOIN Sunday
## 5 5 probs ASSAULT 0.19387537 0.007413903 TENDERLOIN Thursday
## 6 6 probs ASSAULT 0.19424470 0.004026416 TENDERLOIN Tuesday
## 7 7 probs ASSAULT 0.18748667 0.007646783 TENDERLOIN Wednesday
## 8 8 probs ASSAULT 0.16663037 0.005263419 RICHMOND Friday
## 9 9 probs ASSAULT 0.17601746 0.008187888 RICHMOND Monday
## 10 10 probs ASSAULT 0.18093521 0.006584115 RICHMOND Saturday
## 11 11 probs ASSAULT 0.19736750 0.005492126 RICHMOND Sunday
## 12 12 probs ASSAULT 0.16838408 0.006982086 RICHMOND Thursday
## 13 13 probs ASSAULT 0.17069994 0.003170870 RICHMOND Tuesday
## 14 14 probs ASSAULT 0.17099635 0.007372097 RICHMOND Wednesday
## 15 1 probs DRUG/NARCOTIC 0.46581221 0.002597267 TENDERLOIN Friday
## 16 2 probs DRUG/NARCOTIC 0.50014661 0.001959300 TENDERLOIN Monday
## 17 3 probs DRUG/NARCOTIC 0.42007509 0.004293186 TENDERLOIN Saturday
## 18 4 probs DRUG/NARCOTIC 0.42872910 0.003948120 TENDERLOIN Sunday
## 19 5 probs DRUG/NARCOTIC 0.51795000 0.001949593 TENDERLOIN Thursday
## 20 6 probs DRUG/NARCOTIC 0.52086682 0.007446236 TENDERLOIN Tuesday
## 21 7 probs DRUG/NARCOTIC 0.54002868 0.002666268 TENDERLOIN Wednesday
## 22 8 probs DRUG/NARCOTIC 0.04944552 0.003915027 RICHMOND Friday
## 23 9 probs DRUG/NARCOTIC 0.05753044 0.002999987 RICHMOND Monday
## 24 10 probs DRUG/NARCOTIC 0.04243108 0.006053404 RICHMOND Saturday
## 25 11 probs DRUG/NARCOTIC 0.04496576 0.005449351 RICHMOND Sunday
## 26 12 probs DRUG/NARCOTIC 0.06092432 0.003776777 RICHMOND Thursday
## 27 13 probs DRUG/NARCOTIC 0.06199196 0.008108777 RICHMOND Tuesday
## 28 14 probs DRUG/NARCOTIC 0.06670490 0.003813318 RICHMOND Wednesday
## 29 1 probs LARCENY/THEFT 0.29077933 0.007962724 TENDERLOIN Friday
## 30 2 probs LARCENY/THEFT 0.26560692 0.009942019 TENDERLOIN Monday
## 31 3 probs LARCENY/THEFT 0.30606108 0.011494559 TENDERLOIN Saturday
## 32 4 probs LARCENY/THEFT 0.28686823 0.010263234 TENDERLOIN Sunday
## 33 5 probs LARCENY/THEFT 0.26176575 0.008555238 TENDERLOIN Thursday
## 34 6 probs LARCENY/THEFT 0.25933313 0.008657594 TENDERLOIN Tuesday
## 35 7 probs LARCENY/THEFT 0.24794975 0.009270139 TENDERLOIN Wednesday
## 36 8 probs LARCENY/THEFT 0.54666349 0.006314808 RICHMOND Friday
## 37 9 probs LARCENY/THEFT 0.54110340 0.008026769 RICHMOND Monday
## 38 10 probs LARCENY/THEFT 0.54752727 0.007613625 RICHMOND Saturday
## 39 11 probs LARCENY/THEFT 0.53287082 0.007784299 RICHMOND Sunday
## 40 12 probs LARCENY/THEFT 0.54532596 0.009163512 RICHMOND Thursday
## 41 13 probs LARCENY/THEFT 0.54664723 0.005920510 RICHMOND Tuesday
## 42 14 probs LARCENY/THEFT 0.54243172 0.006886299 RICHMOND Wednesday
## 43 1 probs VEHICLE THEFT 0.03080879 0.001798800 TENDERLOIN Friday
## 44 2 probs VEHICLE THEFT 0.02700348 0.002201869 TENDERLOIN Monday
## 45 3 probs VEHICLE THEFT 0.03126405 0.001761442 TENDERLOIN Saturday
## 46 4 probs VEHICLE THEFT 0.02954299 0.002952967 TENDERLOIN Sunday
## 47 5 probs VEHICLE THEFT 0.02640888 0.001026810 TENDERLOIN Thursday
## 48 6 probs VEHICLE THEFT 0.02555535 0.001469659 TENDERLOIN Tuesday
## 49 7 probs VEHICLE THEFT 0.02453490 0.002094417 TENDERLOIN Wednesday
## 50 8 probs VEHICLE THEFT 0.23726062 0.005015415 RICHMOND Friday
## 51 9 probs VEHICLE THEFT 0.22534869 0.006536327 RICHMOND Monday
## 52 10 probs VEHICLE THEFT 0.22910644 0.005736426 RICHMOND Saturday
## 53 11 probs VEHICLE THEFT 0.22479591 0.008327058 RICHMOND Sunday
## 54 12 probs VEHICLE THEFT 0.22536564 0.004681631 RICHMOND Thursday
## 55 13 probs VEHICLE THEFT 0.22066087 0.004907536 RICHMOND Tuesday
## 56 14 probs VEHICLE THEFT 0.21986703 0.005971657 RICHMOND Wednesdayp %>%
select(group, predicted, DayOfWeek, PdDistrict) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 14 × 6
## DayOfWeek PdDistrict ASSAULT `DRUG/NARCOTIC` `LARCENY/THEFT` `VEHICLE THEFT`
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Friday TENDERLOIN 0.213 0.466 0.291 0.0308
## 2 Monday TENDERLOIN 0.207 0.500 0.266 0.0270
## 3 Saturday TENDERLOIN 0.243 0.420 0.306 0.0313
## 4 Sunday TENDERLOIN 0.255 0.429 0.287 0.0295
## 5 Thursday TENDERLOIN 0.194 0.518 0.262 0.0264
## 6 Tuesday TENDERLOIN 0.194 0.521 0.259 0.0256
## 7 Wednesday TENDERLOIN 0.187 0.540 0.248 0.0245
## 8 Friday RICHMOND 0.167 0.0494 0.547 0.237
## 9 Monday RICHMOND 0.176 0.0575 0.541 0.225
## 10 Saturday RICHMOND 0.181 0.0424 0.548 0.229
## 11 Sunday RICHMOND 0.197 0.0450 0.533 0.225
## 12 Thursday RICHMOND 0.168 0.0609 0.545 0.225
## 13 Tuesday RICHMOND 0.171 0.0620 0.547 0.221
## 14 Wednesday RICHMOND 0.171 0.0667 0.542 0.220Richmond is obviously not a drug-dealing kind of place; most of its crimes are theft of one kind or another. But the predicted effect of weekday vs. weekend is the same: Richmond doesn’t have many assaults or drug crimes, but it also has more assaults and fewer drug crimes on the weekend than during the week. There is not much effect of day of the week on the other two crime types in either place.
The consistency of pattern, even though the prevalence of the different crime types differs by location, is a feature of the model: we fitted an additive model, that says there is an effect of weekday, and independently there is an effect of location. The pattern over weekday is the same for each location, implied by the model. This may or may not be supported by the actual data.
The way to assess this is to fit a model with interaction (we will see more of this when we revisit ANOVA later), and compare the fit:
sfcrime.3 <- update(sfcrime.1,.~.+DayOfWeek*PdDistrict)## # weights: 284 (210 variable)
## initial value 498411.639069
## iter 10 value 429631.807781
## iter 20 value 429261.427210
## iter 30 value 428111.625547
## iter 40 value 423807.031450
## iter 50 value 421129.496196
## iter 60 value 420475.833895
## iter 70 value 419523.235916
## iter 80 value 418621.612920
## iter 90 value 418147.629782
## iter 100 value 418036.670485
## final value 418036.670485
## stopped after 100 iterations# anova(sfcrime.1,sfcrime.3)This one didn’t actually complete the fitting process: it got to 100 times around and stopped (since that’s the default limit). We can make it go a bit further thus:
sfcrime.3 <- update(sfcrime.1, .~.+DayOfWeek*PdDistrict, maxit=300)## # weights: 284 (210 variable)
## initial value 498411.639069
## iter 10 value 429631.807781
## iter 20 value 429261.427210
## iter 30 value 428111.625547
## iter 40 value 423807.031450
## iter 50 value 421129.496196
## iter 60 value 420475.833895
## iter 70 value 419523.235916
## iter 80 value 418621.612920
## iter 90 value 418147.629782
## iter 100 value 418036.670485
## iter 110 value 417957.337016
## iter 120 value 417908.465189
## iter 130 value 417890.580843
## iter 140 value 417874.839492
## iter 150 value 417867.449342
## iter 160 value 417862.626213
## iter 170 value 417858.654628
## final value 417858.031854
## convergedanova(sfcrime.1, sfcrime.3)## Likelihood ratio tests of Multinomial Models
##
## Response: Category
## Model Resid. df Resid. Dev Test
## 1 DayOfWeek + PdDistrict 1078536 836300.0
## 2 DayOfWeek + PdDistrict + DayOfWeek:PdDistrict 1078374 835716.1 1 vs 2
## Df LR stat. Pr(Chi)
## 1
## 2 162 583.8955 0This time, we got to the end. (The maxit=300 gets passed on
to multinom, and says “go around up to 300 times if needed”.)
As you will see if you try it, this takes a bit of time to
run.
This anova is also strongly significant, but in the light of
the previous discussion, the differential effect of day of week in
different districts might not be very big. We can even assess that; we
have all the machinery for the predictions, and we just have to apply
them to this model:
p <- predictions(sfcrime.3, newdata = new, type = "probs")
p## rowid type group predicted std.error PdDistrict DayOfWeek
## 1 1 probs ASSAULT 0.21635131 0.02155430 TENDERLOIN Friday
## 2 2 probs ASSAULT 0.20613374 0.02842624 TENDERLOIN Monday
## 3 3 probs ASSAULT 0.23685611 0.02828315 TENDERLOIN Saturday
## 4 4 probs ASSAULT 0.25703306 0.02191080 TENDERLOIN Sunday
## 5 5 probs ASSAULT 0.19518511 0.02517462 TENDERLOIN Thursday
## 6 6 probs ASSAULT 0.19071370 0.02817112 TENDERLOIN Tuesday
## 7 7 probs ASSAULT 0.19063090 0.03962003 TENDERLOIN Wednesday
## 8 8 probs ASSAULT 0.17560619 0.01420215 RICHMOND Friday
## 9 9 probs ASSAULT 0.16688547 0.01644987 RICHMOND Monday
## 10 10 probs ASSAULT 0.18595137 0.02245398 RICHMOND Saturday
## 11 11 probs ASSAULT 0.18514063 0.01688619 RICHMOND Sunday
## 12 12 probs ASSAULT 0.18111853 0.01994638 RICHMOND Thursday
## 13 13 probs ASSAULT 0.17581107 0.01285312 RICHMOND Tuesday
## 14 14 probs ASSAULT 0.16037957 0.03320285 RICHMOND Wednesday
## 15 1 probs DRUG/NARCOTIC 0.44958359 0.01570585 TENDERLOIN Friday
## 16 2 probs DRUG/NARCOTIC 0.51256922 0.01533847 TENDERLOIN Monday
## 17 3 probs DRUG/NARCOTIC 0.41026767 0.02143317 TENDERLOIN Saturday
## 18 4 probs DRUG/NARCOTIC 0.40019234 0.02278750 TENDERLOIN Sunday
## 19 5 probs DRUG/NARCOTIC 0.52867900 0.01331087 TENDERLOIN Thursday
## 20 6 probs DRUG/NARCOTIC 0.53955543 0.04161672 TENDERLOIN Tuesday
## 21 7 probs DRUG/NARCOTIC 0.54577677 0.01464654 TENDERLOIN Wednesday
## 22 8 probs DRUG/NARCOTIC 0.05241934 0.01233050 RICHMOND Friday
## 23 9 probs DRUG/NARCOTIC 0.06038366 0.01233232 RICHMOND Monday
## 24 10 probs DRUG/NARCOTIC 0.04414673 0.01727380 RICHMOND Saturday
## 25 11 probs DRUG/NARCOTIC 0.05211970 0.01494999 RICHMOND Sunday
## 26 12 probs DRUG/NARCOTIC 0.05621201 0.00790321 RICHMOND Thursday
## 27 13 probs DRUG/NARCOTIC 0.05587178 0.02526889 RICHMOND Tuesday
## 28 14 probs DRUG/NARCOTIC 0.06279956 0.01106770 RICHMOND Wednesday
## 29 1 probs LARCENY/THEFT 0.30069679 0.02770219 TENDERLOIN Friday
## 30 2 probs LARCENY/THEFT 0.25699538 0.03607633 TENDERLOIN Monday
## 31 3 probs LARCENY/THEFT 0.32218205 0.02917479 TENDERLOIN Saturday
## 32 4 probs LARCENY/THEFT 0.31011835 0.03264045 TENDERLOIN Sunday
## 33 5 probs LARCENY/THEFT 0.25284647 0.03930132 TENDERLOIN Thursday
## 34 6 probs LARCENY/THEFT 0.24197567 0.04089551 TENDERLOIN Tuesday
## 35 7 probs LARCENY/THEFT 0.24015853 0.04586929 TENDERLOIN Wednesday
## 36 8 probs LARCENY/THEFT 0.54477973 0.01952290 RICHMOND Friday
## 37 9 probs LARCENY/THEFT 0.55376000 0.02353809 RICHMOND Monday
## 38 10 probs LARCENY/THEFT 0.52442616 0.02077298 RICHMOND Saturday
## 39 11 probs LARCENY/THEFT 0.54039667 0.02148531 RICHMOND Sunday
## 40 12 probs LARCENY/THEFT 0.53944839 0.03034502 RICHMOND Thursday
## 41 13 probs LARCENY/THEFT 0.54714052 0.02822038 RICHMOND Tuesday
## 42 14 probs LARCENY/THEFT 0.55255651 0.04051577 RICHMOND Wednesday
## 43 1 probs VEHICLE THEFT 0.03336831 0.02261102 TENDERLOIN Friday
## 44 2 probs VEHICLE THEFT 0.02430166 0.02377108 TENDERLOIN Monday
## 45 3 probs VEHICLE THEFT 0.03069417 0.02558296 TENDERLOIN Saturday
## 46 4 probs VEHICLE THEFT 0.03265625 0.03796627 TENDERLOIN Sunday
## 47 5 probs VEHICLE THEFT 0.02328943 0.02206283 TENDERLOIN Thursday
## 48 6 probs VEHICLE THEFT 0.02775520 0.02650181 TENDERLOIN Tuesday
## 49 7 probs VEHICLE THEFT 0.02343381 0.03557974 TENDERLOIN Wednesday
## 50 8 probs VEHICLE THEFT 0.22719474 0.02162989 RICHMOND Friday
## 51 9 probs VEHICLE THEFT 0.21897087 0.01991857 RICHMOND Monday
## 52 10 probs VEHICLE THEFT 0.24547574 0.02686280 RICHMOND Saturday
## 53 11 probs VEHICLE THEFT 0.22234300 0.03436623 RICHMOND Sunday
## 54 12 probs VEHICLE THEFT 0.22322107 0.02014785 RICHMOND Thursday
## 55 13 probs VEHICLE THEFT 0.22117663 0.02722655 RICHMOND Tuesday
## 56 14 probs VEHICLE THEFT 0.22426437 0.01586793 RICHMOND Wednesdayp %>%
select(group, predicted, DayOfWeek, PdDistrict) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 14 × 6
## DayOfWeek PdDistrict ASSAULT `DRUG/NARCOTIC` `LARCENY/THEFT` `VEHICLE THEFT`
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Friday TENDERLOIN 0.216 0.450 0.301 0.0334
## 2 Monday TENDERLOIN 0.206 0.513 0.257 0.0243
## 3 Saturday TENDERLOIN 0.237 0.410 0.322 0.0307
## 4 Sunday TENDERLOIN 0.257 0.400 0.310 0.0327
## 5 Thursday TENDERLOIN 0.195 0.529 0.253 0.0233
## 6 Tuesday TENDERLOIN 0.191 0.540 0.242 0.0278
## 7 Wednesday TENDERLOIN 0.191 0.546 0.240 0.0234
## 8 Friday RICHMOND 0.176 0.0524 0.545 0.227
## 9 Monday RICHMOND 0.167 0.0604 0.554 0.219
## 10 Saturday RICHMOND 0.186 0.0441 0.524 0.245
## 11 Sunday RICHMOND 0.185 0.0521 0.540 0.222
## 12 Thursday RICHMOND 0.181 0.0562 0.539 0.223
## 13 Tuesday RICHMOND 0.176 0.0559 0.547 0.221
## 14 Wednesday RICHMOND 0.160 0.0628 0.553 0.224It doesn’t look much different. Maybe the Tenderloin has a larger weekend increase in assaults than Richmond does.
\(\blacksquare\)
28.11 Crimes in San Francisco – the data
The data in link is a huge dataset of crimes committed in San Francisco between 2003 and 2015. The variables are:
Dates: the date and time of the crimeCategory: the category of crime, eg. “larceny” or “vandalism” (response).Descript: detailed description of crime.DayOfWeek: the day of the week of the crime.PdDistrict: the name of the San Francisco Police Department district in which the crime took place.Resolution: how the crime was resolvedAddress: approximate street address of crimeX: longitudeY: latitude
Our aim is to see whether the category of crime depends on the day of the week and the district in which it occurred. However, there are a lot of crime categories, so we will focus on the top four “interesting” ones, which we will have to discover.
- Read in the data and verify that you have these columns and a lot of rows. (The data may take a moment to read in. You will see why.)
Solution
my_url="http://utsc.utoronto.ca/~butler/d29/sfcrime.csv"
sfcrime=read_csv(my_url)## Rows: 878049 Columns: 9
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (6): Category, Descript, DayOfWeek, PdDistrict, Resolution, Address
## dbl (2): X, Y
## dttm (1): Dates
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.sfcrime## # A tibble: 878,049 × 9
## Dates Category Descript DayOfWeek PdDistrict Resolution Address
## <dttm> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 2015-05-13 23:53:00 WARRANTS WARRANT… Wednesday NORTHERN ARREST, B… OAK ST…
## 2 2015-05-13 23:53:00 OTHER O… TRAFFIC… Wednesday NORTHERN ARREST, B… OAK ST…
## 3 2015-05-13 23:33:00 OTHER O… TRAFFIC… Wednesday NORTHERN ARREST, B… VANNES…
## 4 2015-05-13 23:30:00 LARCENY… GRAND T… Wednesday NORTHERN NONE 1500 B…
## 5 2015-05-13 23:30:00 LARCENY… GRAND T… Wednesday PARK NONE 100 Bl…
## 6 2015-05-13 23:30:00 LARCENY… GRAND T… Wednesday INGLESIDE NONE 0 Bloc…
## 7 2015-05-13 23:30:00 VEHICLE… STOLEN … Wednesday INGLESIDE NONE AVALON…
## 8 2015-05-13 23:30:00 VEHICLE… STOLEN … Wednesday BAYVIEW NONE KIRKWO…
## 9 2015-05-13 23:00:00 LARCENY… GRAND T… Wednesday RICHMOND NONE 600 Bl…
## 10 2015-05-13 23:00:00 LARCENY… GRAND T… Wednesday CENTRAL NONE JEFFER…
## # … with 878,039 more rows, and 2 more variables: X <dbl>, Y <dbl>Those columns indeed, and pushing a million rows! That’s why it took so long!
There are also 39 categories of crime, so we need to cut that down some. There are only ten districts, however, so we should be able to use that as is.
\(\blacksquare\)
- How is the response variable here different to the one in
the question about steak preferences (and therefore why would
multinomfrom packagennetbe the method of choice)?
Solution
Steak preferences have a natural order, while crime categories do
not. Since they are unordered, multinom is better than
polr.
\(\blacksquare\)
- Find out which crime categories there are, and arrange them in order of how many crimes there were in each category.
Solution
This can be the one you know, a group-by and summarize, followed
by arrange to sort:
sfcrime %>% group_by(Category) %>%
summarize(count=n()) %>%
arrange(desc(count))## # A tibble: 39 × 2
## Category count
## <chr> <int>
## 1 LARCENY/THEFT 174900
## 2 OTHER OFFENSES 126182
## 3 NON-CRIMINAL 92304
## 4 ASSAULT 76876
## 5 DRUG/NARCOTIC 53971
## 6 VEHICLE THEFT 53781
## 7 VANDALISM 44725
## 8 WARRANTS 42214
## 9 BURGLARY 36755
## 10 SUSPICIOUS OCC 31414
## # … with 29 more rowsor this one does the same thing and saves a step:
sfcrime %>% count(Category) %>%
arrange(desc(n))## # A tibble: 39 × 2
## Category n
## <chr> <int>
## 1 LARCENY/THEFT 174900
## 2 OTHER OFFENSES 126182
## 3 NON-CRIMINAL 92304
## 4 ASSAULT 76876
## 5 DRUG/NARCOTIC 53971
## 6 VEHICLE THEFT 53781
## 7 VANDALISM 44725
## 8 WARRANTS 42214
## 9 BURGLARY 36755
## 10 SUSPICIOUS OCC 31414
## # … with 29 more rowsFor this one, do the count step first, to see what you
get. It produces a two-column data frame with the column of counts
called n. So now you know that the second line has to be
arrange(desc(n)), whereas before you tried count,
all you knew is that it was arrange-desc-something.
You need to sort in descending order so that the categories you want to see actually do appear at the top.
\(\blacksquare\)
- Which are the four most frequent “interesting” crime
categories, that is to say, not including “other offenses” and
“non-criminal”? Get them into a vector called
my.crimes. See if you can find a way of doing this that doesn’t involve typing them in (for full marks).
Solution
The most frequent interesting ones are, in order, larceny-theft,
assault, drug-narcotic and vehicle theft.
The fact that “other offenses” is so big indicates that there
are a lot of possible crimes out there, and even 39 categories of
crime isn’t enough. “Non-criminal” means, I think, that the
police were called, but on arriving at the scene, they found that
no law had been broken.
I think the easy way to get the “top four” crimes out is to pull
them out of the data frame that count produces. They are
rows 1, 4, 5 and 6, so add a slice to your pipeline:
my.rows=c(1,4,5,6)
my.crimes = sfcrime %>% count(Category) %>%
arrange(desc(n)) %>%
slice(my.rows) %>% pull(Category)
my.crimes## [1] "LARCENY/THEFT" "ASSAULT" "DRUG/NARCOTIC" "VEHICLE THEFT"I just want the Category column (as a vector), and
pull is the way to get that. (If I don’t do pull, I
get a data frame.)
If you can’t think of anything, just type them into a vector with
c, or better, copy-paste them from the console. But that’s a
last resort, and would cost you a point. If you do this, they have to
match exactly, UPPERCASE and all.
\(\blacksquare\)
- (Digression, but needed for the next part.) The R vector
letterscontains the lowercase letters fromatoz. Consider the vector('a','m',3,'Q'). Some of these are found amongst the lowercase letters, and some not. Type these into a vectorvand explain briefly whyv %in% lettersproduces what it does.
Solution
This is the ultimate “try it and see”:
v=c('a','m',3,'Q')
v %in% letters## [1] TRUE TRUE FALSE FALSEThe first two elements of the answer are TRUE because
lowercase-a and lowercase-m can be found in the lowercase letters
somewhere. The other two are false because the number 3 and the
uppercase-Q cannot be found anywhere in the lowercase letters.
The name is %in% because it’s asking whether each element of
the first vector (one at a time) is in the set defined by the
second thing: “is a a lowercase letter?” …
is “Q a lowercase letter?”
and getting the answers “yes, yes, no, no”.
\(\blacksquare\)
- We are going to
filteronly the rows of our data frame that have one of the crimes inmy.crimesas theirCategory. Also,selectonly the columnsCategory,DayOfWeekandPdDistrict. Save the resulting data frame and display its structure. (You should have a lot fewer rows than you did before.)
Solution
The hard part about this is to get the inputs to %in% the
right way around. We are testing the things in Category one
at a time for membership in the set in my.crimes, so this:
sfcrimea = sfcrime %>% filter(Category %in% my.crimes) %>%
select(c(Category,DayOfWeek,PdDistrict))
sfcrimea## # A tibble: 359,528 × 3
## Category DayOfWeek PdDistrict
## <chr> <chr> <chr>
## 1 LARCENY/THEFT Wednesday NORTHERN
## 2 LARCENY/THEFT Wednesday PARK
## 3 LARCENY/THEFT Wednesday INGLESIDE
## 4 VEHICLE THEFT Wednesday INGLESIDE
## 5 VEHICLE THEFT Wednesday BAYVIEW
## 6 LARCENY/THEFT Wednesday RICHMOND
## 7 LARCENY/THEFT Wednesday CENTRAL
## 8 LARCENY/THEFT Wednesday CENTRAL
## 9 LARCENY/THEFT Wednesday NORTHERN
## 10 ASSAULT Wednesday INGLESIDE
## # … with 359,518 more rowsI had trouble thinking of a good name for this one, so I put an “a” on the end. (I would have used a number, but I prefer those for models.)
Down to a “mere” 359,000 rows. Don’t be stressed that the
Category factor still has 39 levels (the original 39 crime
categories); only four of them have any data in them:
sfcrimea %>% count(Category)## # A tibble: 4 × 2
## Category n
## <chr> <int>
## 1 ASSAULT 76876
## 2 DRUG/NARCOTIC 53971
## 3 LARCENY/THEFT 174900
## 4 VEHICLE THEFT 53781So all of the crimes that are left are one of the four Categories we want to look at.
\(\blacksquare\)
- Save these data in a file
sfcrime1.csv.
Solution
This is write_csv again:
write_csv(sfcrimea,"sfcrime1.csv")\(\blacksquare\)
28.12 What sports do these athletes play?
The data at link are physical and physiological measurements of 202 male and female Australian elite athletes. The data values are separated by tabs. We are going to see whether we can predict the sport an athlete plays from their height and weight.
The sports, if you care, are respectively basketball, “field athletics” (eg. shot put, javelin throw, long jump etc.), gymnastics, netball, rowing, swimming, 400m running, tennis, sprinting (100m or 200m running), water polo.
- Read in the data and display the first few rows.
Solution
The data values are separated by tabs, so read_tsv is
the thing:
my_url <- "http://ritsokiguess.site/datafiles/ais.txt"
athletes <- read_tsv(my_url)## Rows: 202 Columns: 13
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## chr (2): Sex, Sport
## dbl (11): RCC, WCC, Hc, Hg, Ferr, BMI, SSF, %Bfat, LBM, Ht, Wt
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.athletes## # A tibble: 202 × 13
## Sex Sport RCC WCC Hc Hg Ferr BMI SSF `%Bfat` LBM Ht
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 female Netball 4.56 13.3 42.2 13.6 20 19.2 49 11.3 53.1 177.
## 2 female Netball 4.15 6 38 12.7 59 21.2 110. 25.3 47.1 173.
## 3 female Netball 4.16 7.6 37.5 12.3 22 21.4 89 19.4 53.4 176
## 4 female Netball 4.32 6.4 37.7 12.3 30 21.0 98.3 19.6 48.8 170.
## 5 female Netball 4.06 5.8 38.7 12.8 78 21.8 122. 23.1 56.0 183
## 6 female Netball 4.12 6.1 36.6 11.8 21 21.4 90.4 16.9 56.4 178.
## 7 female Netball 4.17 5 37.4 12.7 109 21.5 107. 21.3 53.1 177.
## 8 female Netball 3.8 6.6 36.5 12.4 102 24.4 157. 26.6 54.4 174.
## 9 female Netball 3.96 5.5 36.3 12.4 71 22.6 101. 17.9 56.0 174.
## 10 female Netball 4.44 9.7 41.4 14.1 64 22.8 126. 25.0 51.6 174.
## # … with 192 more rows, and 1 more variable: Wt <dbl>If you didn’t remember that, this also works:
athletes <- read_delim(my_url, "\t")## Rows: 202 Columns: 13
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## chr (2): Sex, Sport
## dbl (11): RCC, WCC, Hc, Hg, Ferr, BMI, SSF, %Bfat, LBM, Ht, Wt
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.(this is the R way of expressing “tab”.)
\(\blacksquare\)
- Make a scatterplot of height vs. weight, with the points coloured by what sport the athlete plays. Put height on the \(x\)-axis and weight on the \(y\)-axis.
Solution
I’m doing this to give you a little intuition for later:
ggplot(athletes, aes(x = Ht, y = Wt, colour = Sport)) + geom_point()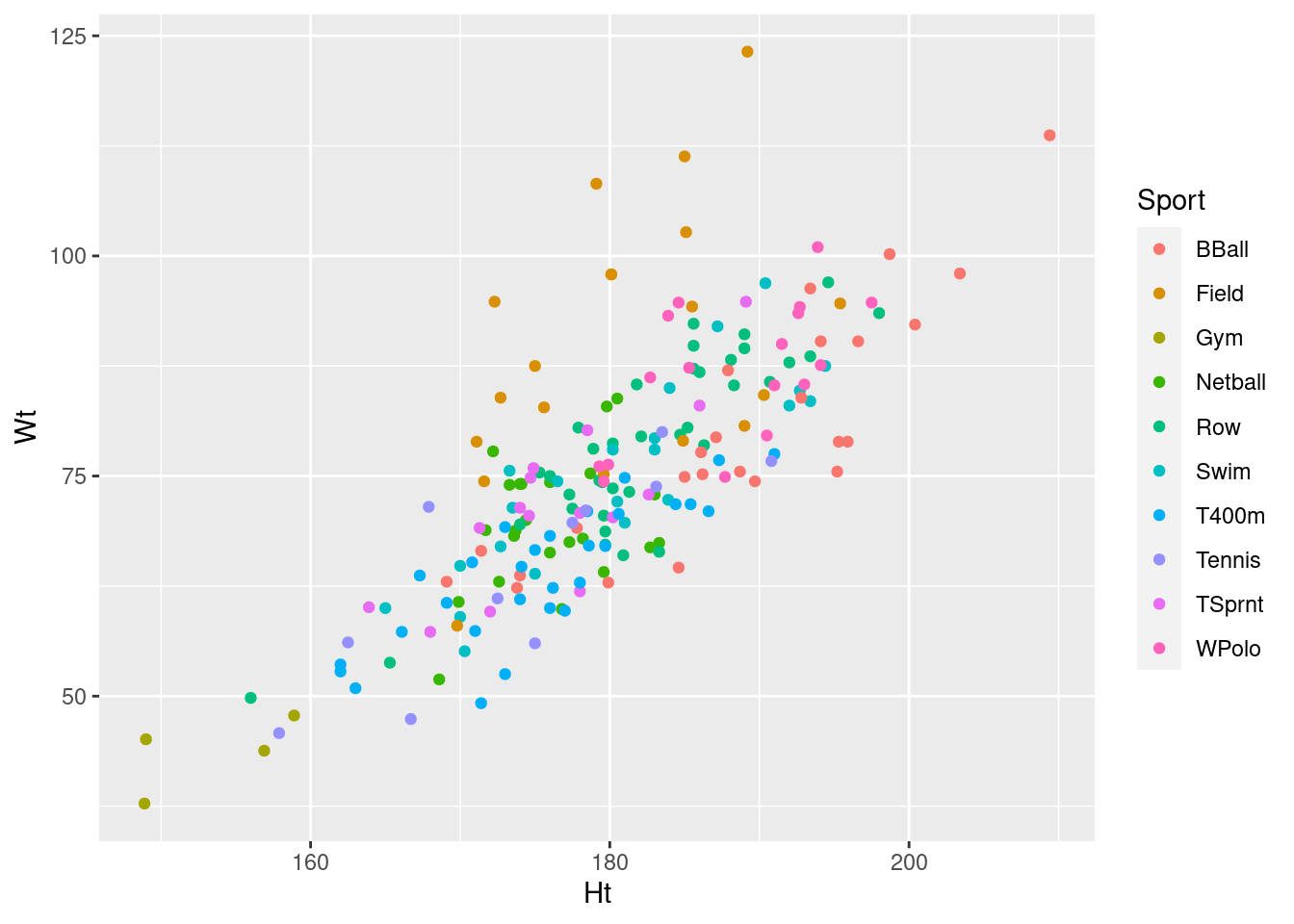
The reason for giving you the axes to use is (i) neither variable is really a response, so it doesn’t actually matter which one goes on which axis, and (ii) I wanted to give the grader something consistent to look at.
\(\blacksquare\)
- Explain briefly why a multinomial model (
multinomfromnnet) would be the best thing to use to predict sport played from the other variables.
Solution
The categories of Sport are not in any kind of order, and
there are more than two of them.
That’s really all you needed, for which two marks was kind of
generous.
\(\blacksquare\)
- Fit a suitable model for predicting sport played from
height and weight. (You don’t need to look at the results.) 100
steps isn’t quite enough, so set
maxitequal to a larger number to allow the estimation to finish.
Solution
120 steps is actually enough, but any number larger than 110 is fine. It doesn’t matter if your guess is way too high. Like this:
library(nnet)
sport.1 <- multinom(Sport ~ Ht + Wt, data = athletes, maxit = 200)## # weights: 40 (27 variable)
## initial value 465.122189
## iter 10 value 410.089598
## iter 20 value 391.426721
## iter 30 value 365.829150
## iter 40 value 355.326457
## iter 50 value 351.255395
## iter 60 value 350.876479
## iter 70 value 350.729699
## iter 80 value 350.532323
## iter 90 value 350.480130
## iter 100 value 350.349271
## iter 110 value 350.312029
## final value 350.311949
## convergedAs long as you see the word converged at the end, you’re
good.
\(\blacksquare\)
- Demonstrate using
anovathatWtshould not be removed from this model.
Solution
The idea is to fit a model without Wt, and then show that
it fits significantly worse. This converges in less than 100 iterations, so you can have maxit or not as you prefer:
sport.2 <- update(sport.1, . ~ . - Wt)## # weights: 30 (18 variable)
## initial value 465.122189
## iter 10 value 447.375728
## iter 20 value 413.597441
## iter 30 value 396.685596
## iter 40 value 394.121380
## iter 50 value 394.116993
## iter 60 value 394.116434
## final value 394.116429
## convergedanova(sport.2, sport.1, test = "Chisq")## Likelihood ratio tests of Multinomial Models
##
## Response: Sport
## Model Resid. df Resid. Dev Test Df LR stat. Pr(Chi)
## 1 Ht 1800 788.2329
## 2 Ht + Wt 1791 700.6239 1 vs 2 9 87.60896 4.884981e-15The P-value is very small indeed, so the bigger model sport.1
is definitely better (or the smaller model sport.2 is
significantly worse, however you want to say it). So taking
Wt out is definitely a mistake.
This is what I would have guessed (I actually wrote the question in anticipation of this being the answer) because weight certainly seems to help in distinguishing the sports. For example, the field athletes seem to be heavy for their height compared to the other athletes (look back at the graph you made).
drop1, the obvious thing, doesn’t work here:
drop1(sport.1, test = "Chisq", trace = T)## trying - Ht## Error in if (trace) {: argument is not interpretable as logicalI gotta figure out what that error is.
Does step?
step(sport.1, direction = "backward", test = "Chisq")## Start: AIC=754.62
## Sport ~ Ht + Wt
##
## trying - Ht
## # weights: 30 (18 variable)
## initial value 465.122189
## iter 10 value 441.367394
## iter 20 value 381.021649
## iter 30 value 380.326030
## final value 380.305003
## converged
## trying - Wt
## # weights: 30 (18 variable)
## initial value 465.122189
## iter 10 value 447.375728
## iter 20 value 413.597441
## iter 30 value 396.685596
## iter 40 value 394.121380
## iter 50 value 394.116993
## iter 60 value 394.116434
## final value 394.116429
## converged
## Df AIC
## <none> 27 754.6239
## - Ht 18 796.6100
## - Wt 18 824.2329## Call:
## multinom(formula = Sport ~ Ht + Wt, data = athletes, maxit = 200)
##
## Coefficients:
## (Intercept) Ht Wt
## Field 59.98535 -0.4671650 0.31466413
## Gym 112.49889 -0.5027056 -0.57087657
## Netball 47.70209 -0.2947852 0.07992763
## Row 35.90829 -0.2517942 0.14164007
## Swim 36.82832 -0.2444077 0.10544986
## T400m 32.73554 -0.1482589 -0.07830622
## Tennis 41.92855 -0.2278949 -0.01979877
## TSprnt 51.43723 -0.3359534 0.12378285
## WPolo 23.35291 -0.2089807 0.18819526
##
## Residual Deviance: 700.6239
## AIC: 754.6239Curiously enough, it does. The final model is the same as the initial one, telling us that neither variable should be removed.
\(\blacksquare\)
- Make a data frame consisting of all combinations of
Ht160, 180 and 200 (cm), andWt50, 75, and 100 (kg), and use it to obtain predicted probabilities of athletes of those heights and weights playing each of the sports. Display the results. You might have to display them smaller, or reduce the number of decimal places51 to fit them on the page.
Solution
To get all combinations of those heights and weights, use datagrid:
new <- datagrid(model = sport.1, Ht = c(160, 180, 200), Wt = c(50, 75, 100))
new## Ht Wt
## 1: 160 50
## 2: 160 75
## 3: 160 100
## 4: 180 50
## 5: 180 75
## 6: 180 100
## 7: 200 50
## 8: 200 75
## 9: 200 100(check: \(3 \times 3 = 9\) rows.)
Then predict:
p <- predictions(sport.1, newdata = new, type = "probs")
p## rowid type group predicted std.error Ht Wt
## 1 1 probs BBall 2.147109e-03 0 160 50
## 2 2 probs BBall 1.044581e-04 0 160 75
## 3 3 probs BBall 5.541050e-08 0 160 100
## 4 4 probs BBall 9.142760e-02 0 180 50
## 5 5 probs BBall 7.787300e-02 0 180 75
## 6 6 probs BBall 5.379479e-04 0 180 100
## 7 7 probs BBall 6.907544e-01 0 200 50
## 8 8 probs BBall 8.889269e-01 0 200 75
## 9 9 probs BBall 2.738448e-01 0 200 100
## 10 1 probs Field 5.676191e-03 0 160 50
## 11 2 probs Field 7.203893e-01 0 160 75
## 12 3 probs Field 9.968725e-01 0 160 100
## 13 4 probs Field 2.116103e-05 0 180 50
## 14 5 probs Field 4.701849e-02 0 180 75
## 15 6 probs Field 8.473139e-01 0 180 100
## 16 7 probs Field 1.399715e-08 0 200 50
## 17 8 probs Field 4.698989e-05 0 200 75
## 18 9 probs Field 3.776290e-02 0 200 100
## 19 1 probs Gym 7.269646e-02 0 160 50
## 20 2 probs Gym 2.240720e-09 0 160 75
## 21 3 probs Gym 7.530504e-19 0 160 100
## 22 4 probs Gym 1.331347e-04 0 180 50
## 23 5 probs Gym 7.184344e-11 0 180 75
## 24 6 probs Gym 3.144321e-19 0 180 100
## 25 7 probs Gym 4.326057e-08 0 200 50
## 26 8 probs Gym 3.527126e-14 0 200 75
## 27 9 probs Gym 6.884080e-21 0 200 100
## 28 1 probs Netball 1.997276e-01 0 160 50
## 29 2 probs Netball 7.166865e-02 0 160 75
## 30 3 probs Netball 2.804029e-04 0 160 100
## 31 4 probs Netball 2.339857e-02 0 180 50
## 32 5 probs Netball 1.469948e-01 0 180 75
## 33 6 probs Netball 7.489597e-03 0 180 100
## 34 7 probs Netball 4.863664e-04 0 200 50
## 35 8 probs Netball 4.616460e-03 0 200 75
## 36 9 probs Netball 1.048940e-02 0 200 100
## 37 1 probs Row 3.205118e-02 0 160 50
## 38 2 probs Row 5.379841e-02 0 160 75
## 39 3 probs Row 9.845934e-04 0 160 100
## 40 4 probs Row 8.871768e-03 0 180 50
## 41 5 probs Row 2.607097e-01 0 180 75
## 42 6 probs Row 6.213668e-02 0 180 100
## 43 7 probs Row 4.357120e-04 0 200 50
## 44 8 probs Row 1.934547e-02 0 200 75
## 45 9 probs Row 2.056153e-01 0 200 100
## 46 1 probs Swim 4.293546e-02 0 160 50
## 47 2 probs Swim 2.916160e-02 0 160 75
## 48 3 probs Swim 2.159577e-04 0 160 100
## 49 4 probs Swim 1.377656e-02 0 180 50
## 50 5 probs Swim 1.638165e-01 0 180 75
## 51 6 probs Swim 1.579859e-02 0 180 100
## 52 7 probs Swim 7.843113e-04 0 200 50
## 53 8 probs Swim 1.409088e-02 0 200 75
## 54 9 probs Swim 6.060159e-02 0 200 100
## 55 1 probs T400m 3.517558e-01 0 160 50
## 56 2 probs T400m 2.416185e-03 0 160 75
## 57 3 probs T400m 1.809594e-07 0 160 100
## 58 4 probs T400m 7.721547e-01 0 180 50
## 59 5 probs T400m 9.285706e-02 0 180 75
## 60 6 probs T400m 9.056685e-05 0 180 100
## 61 7 probs T400m 3.007396e-01 0 200 50
## 62 8 probs T400m 5.464293e-02 0 200 75
## 63 9 probs T400m 2.376695e-03 0 200 100
## 64 1 probs Tennis 1.885847e-01 0 160 50
## 65 2 probs Tennis 5.592833e-03 0 160 75
## 66 3 probs Tennis 1.808504e-06 0 160 100
## 67 4 probs Tennis 8.418976e-02 0 180 50
## 68 5 probs Tennis 4.371258e-02 0 180 75
## 69 6 probs Tennis 1.840761e-04 0 180 100
## 70 7 probs Tennis 6.668610e-03 0 200 50
## 71 8 probs Tennis 5.231368e-03 0 200 75
## 72 9 probs Tennis 9.824069e-04 0 200 100
## 73 1 probs TSprnt 1.033315e-01 0 160 50
## 74 2 probs TSprnt 1.109879e-01 0 160 75
## 75 3 probs TSprnt 1.299813e-03 0 160 100
## 76 4 probs TSprnt 5.313751e-03 0 180 50
## 77 5 probs TSprnt 9.992310e-02 0 180 75
## 78 6 probs TSprnt 1.523963e-02 0 180 100
## 79 7 probs TSprnt 4.848339e-05 0 200 50
## 80 8 probs TSprnt 1.377496e-03 0 200 75
## 81 9 probs TSprnt 9.368804e-03 0 200 100
## 82 1 probs WPolo 1.094045e-03 0 160 50
## 83 2 probs WPolo 5.880674e-03 0 160 75
## 84 3 probs WPolo 3.446523e-04 0 160 100
## 85 4 probs WPolo 7.129763e-04 0 180 50
## 86 5 probs WPolo 6.709478e-02 0 180 75
## 87 6 probs WPolo 5.120898e-02 0 180 100
## 88 7 probs WPolo 8.244005e-05 0 200 50
## 89 8 probs WPolo 1.172154e-02 0 200 75
## 90 9 probs WPolo 3.989581e-01 0 200 100I saved these, because I want to talk about what to do next. The normal procedure is to say that there is a prediction for each group (sport), so we want to grab only the columns we need and pivot wider. But, this time there are 10 sports (see how there are \(9 \times 10 = 90\) rows, with a predicted probability for each height-weight combo for each sport). I suggested reducing the number of decimal places in the predictions; the time to do that is now, while they are all in one column (rather than trying to round a bunch of columns all at once, which is doable but more work). Hence:
p %>%
mutate(predicted = round(predicted, 2)) %>%
select(group, predicted, Ht, Wt) %>%
pivot_wider(names_from = group, values_from = predicted)## # A tibble: 9 × 12
## Ht Wt BBall Field Gym Netball Row Swim T400m Tennis TSprnt WPolo
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 160 50 0 0.01 0.07 0.2 0.03 0.04 0.35 0.19 0.1 0
## 2 160 75 0 0.72 0 0.07 0.05 0.03 0 0.01 0.11 0.01
## 3 160 100 0 1 0 0 0 0 0 0 0 0
## 4 180 50 0.09 0 0 0.02 0.01 0.01 0.77 0.08 0.01 0
## 5 180 75 0.08 0.05 0 0.15 0.26 0.16 0.09 0.04 0.1 0.07
## 6 180 100 0 0.85 0 0.01 0.06 0.02 0 0 0.02 0.05
## 7 200 50 0.69 0 0 0 0 0 0.3 0.01 0 0
## 8 200 75 0.89 0 0 0 0.02 0.01 0.05 0.01 0 0.01
## 9 200 100 0.27 0.04 0 0.01 0.21 0.06 0 0 0.01 0.4This works (although even then it might not all fit onto the page and you’ll have to scroll left and right to see everything).
If you forget to round until the end, you’ll have to do something like this:
p %>%
select(group, predicted, Ht, Wt) %>%
pivot_wider(names_from = group, values_from = predicted) %>%
mutate(across(BBall:WPolo, ~round(., 2)))## # A tibble: 9 × 12
## Ht Wt BBall Field Gym Netball Row Swim T400m Tennis TSprnt WPolo
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 160 50 0 0.01 0.07 0.2 0.03 0.04 0.35 0.19 0.1 0
## 2 160 75 0 0.72 0 0.07 0.05 0.03 0 0.01 0.11 0.01
## 3 160 100 0 1 0 0 0 0 0 0 0 0
## 4 180 50 0.09 0 0 0.02 0.01 0.01 0.77 0.08 0.01 0
## 5 180 75 0.08 0.05 0 0.15 0.26 0.16 0.09 0.04 0.1 0.07
## 6 180 100 0 0.85 0 0.01 0.06 0.02 0 0 0.02 0.05
## 7 200 50 0.69 0 0 0 0 0 0.3 0.01 0 0
## 8 200 75 0.89 0 0 0 0.02 0.01 0.05 0.01 0 0.01
## 9 200 100 0.27 0.04 0 0.01 0.21 0.06 0 0 0.01 0.4In words, “for each column from BBall through WPolo, replace it by itself rounded to 2 decimals”.
There’s nothing magic about two decimals; three or maybe even four would be fine. A small enough number that you can see most or all of the columns at once, and easily compare the numbers for size (which is hard when some of them are in scientific notation).
Extra: you can even abbreviate the sport names, like this:
p %>%
mutate(predicted = round(predicted, 2),
group = abbreviate(group, 3)) %>%
select(group, predicted, Ht, Wt) %>%
pivot_wider(names_from = group, values_from = predicted) ## # A tibble: 9 × 12
## Ht Wt BBl Fld Gym Ntb Row Swm T40 Tnn TSp WPl
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 160 50 0 0.01 0.07 0.2 0.03 0.04 0.35 0.19 0.1 0
## 2 160 75 0 0.72 0 0.07 0.05 0.03 0 0.01 0.11 0.01
## 3 160 100 0 1 0 0 0 0 0 0 0 0
## 4 180 50 0.09 0 0 0.02 0.01 0.01 0.77 0.08 0.01 0
## 5 180 75 0.08 0.05 0 0.15 0.26 0.16 0.09 0.04 0.1 0.07
## 6 180 100 0 0.85 0 0.01 0.06 0.02 0 0 0.02 0.05
## 7 200 50 0.69 0 0 0 0 0 0.3 0.01 0 0
## 8 200 75 0.89 0 0 0 0.02 0.01 0.05 0.01 0 0.01
## 9 200 100 0.27 0.04 0 0.01 0.21 0.06 0 0 0.01 0.4Once again, the time to do this is while everything is long, so that you only have one column to work with.52 Again, you can do this at the end, but it’s more work, because you are now renaming columns:
p %>%
mutate(predicted = round(predicted, 2)) %>%
select(group, predicted, Ht, Wt) %>%
pivot_wider(names_from = group, values_from = predicted) %>%
rename_with(~abbreviate(., 3), everything())## # A tibble: 9 × 12
## Ht Wt BBl Fld Gym Ntb Row Swm T40 Tnn TSp WPl
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 160 50 0 0.01 0.07 0.2 0.03 0.04 0.35 0.19 0.1 0
## 2 160 75 0 0.72 0 0.07 0.05 0.03 0 0.01 0.11 0.01
## 3 160 100 0 1 0 0 0 0 0 0 0 0
## 4 180 50 0.09 0 0 0.02 0.01 0.01 0.77 0.08 0.01 0
## 5 180 75 0.08 0.05 0 0.15 0.26 0.16 0.09 0.04 0.1 0.07
## 6 180 100 0 0.85 0 0.01 0.06 0.02 0 0 0.02 0.05
## 7 200 50 0.69 0 0 0 0 0 0.3 0.01 0 0
## 8 200 75 0.89 0 0 0 0.02 0.01 0.05 0.01 0 0.01
## 9 200 100 0.27 0.04 0 0.01 0.21 0.06 0 0 0.01 0.4rename doesn’t work with across (you will get an error if you try it), so you need to use rename_with53. This has syntax “how to rename” first (here, with an abbreviated version of itself), and “what to rename” second (here, everything, or BBall through WPolo if you prefer).
\(\blacksquare\)
- For an athlete who is 180 cm tall and weighs 100 kg, what sport would you guess they play? How sure are you that you are right? Explain briefly.
Solution
Find this height and weight in your predictions (it’s row 6). Look
along the line for the highest probability, which is 0.85 for
Field (that is, field athletics). All the other
probabilities are much smaller (the biggest of the others is
0.06). So this means we would guess the athlete to be a field
athlete, and because the predicted probability is so big, we are
very likely to be right.
This kind of thought process is characteristic of discriminant
analysis, which we’ll see more of later in the course.
Compare that with the scatterplot you drew earlier: the field
athletes do seem to be distinct from the rest in terms of
height-weight combination.
Some of the other height-weight combinations are almost equally
obvious: for example, very tall people who are not very heavy are
likely to play basketball. 400m runners are likely to be of
moderate height but light weight. Some of the other sports, or
height-weight combinations, are difficult to judge. Consider also
that we have mixed up males and females in this data set. We might
gain some clarity by including Sex in the model, and also
in the predictions. But I wanted to keep this question relatively
simple for you, and I wanted to stop from getting unreasonably
long. (You can decide whether you think it’s already too long.)
\(\blacksquare\)
For this, use
round.↩︎If you do not take out the
NAvalues, they are shown separately on the end of thesummaryfor that column.↩︎The American Green party is more libertarian than Green parties elsewhere.↩︎
Northern Ireland’s political parties distinguish themselves by politics and religion. Northern Ireland has always had political tensions between its Protestants and its Catholics.↩︎
There are three political parties; using the first as a baseline, there are therefore \(3-1=2\) coefficients for each variable.↩︎
It amuses me that Toronto’s current (2021) mayor, named Tory, is politically a Tory.↩︎
When you have variables that are categories, you might have more than one individual with exactly the same categories; on the other hand, if they had measured Size as, say, length in centimetres, it would have been very unlikely to get two alligators of exactly the same size.↩︎
Discovered by me two minutes ago.↩︎
Just don’t use a measure of spread, which will give you zero.↩︎
There were only 216 alligators total, which is a small sample size for this kind of thing, especially with all those parameters to estimate.↩︎
Better, because it doesn’t do predictions for all the other districts as well, only to throw them away, which is what we did above. When the predictions take so long, it’s worth trying to speed it up.↩︎
I know I just said not to do this kind of thing, but counting is really fast, while unnecessary predictions are really slow.↩︎
There is only one district, so we can just put that in here.↩︎
The same thing applies if you are doing something like webscraping, that is to say downloading stuff from the web that you then want to do something with. Download it once and save it, then you can take as long as you need to decide what you’re doing with it.↩︎
Statistical significance as an idea grew up in the days before “big data”.↩︎
For this, use
round.↩︎abbreviatecomes from base R; it returns an abbreviated version of its (text) input, with a minimum length of its second input. If two abbreviations come out the same, it makes them longer until they are different. It has some heuristics to get things short enough: remove vowels, then remove letters on the end.↩︎Which I learned about today.↩︎